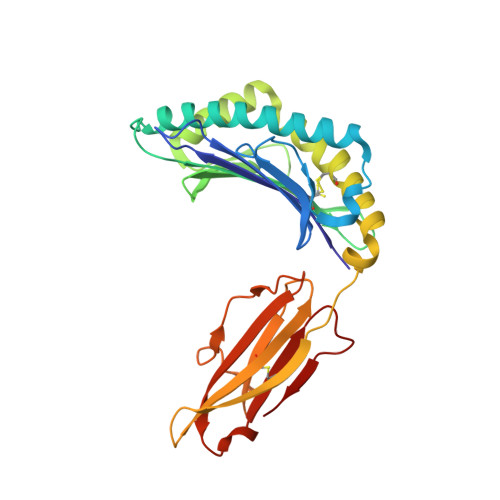
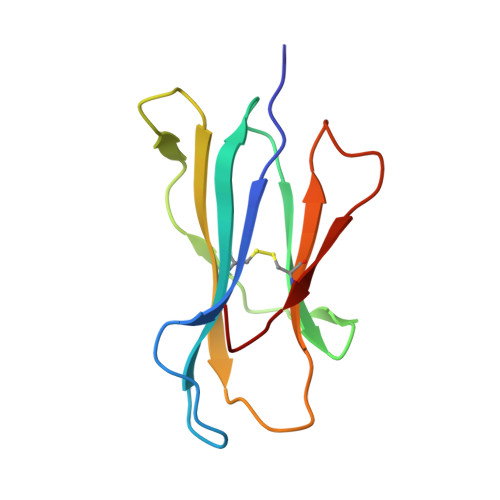
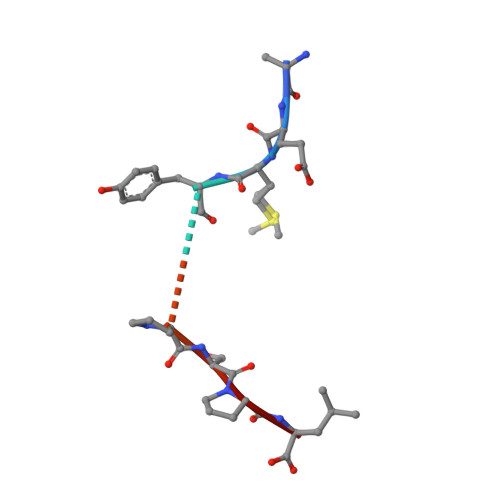
The impact of human leukocyte antigen (HLA) micropolymorphism on ligand specificity within the HLA-B*41 allotypic family
Bade-Doding, C., Theodossis, A., Gras, S., Kjer-Nielsen, L., Eiz-Vesper, B., Seltsam, A., Huyton, T., Rossjohn, J., McCluskey, J., Blasczyk, R.(2011) Haematologica 96: 110-118
- PubMed: 20934997
- DOI: https://doi.org/10.3324/haematol.2010.030924
- Primary Citation of Related Structures:
3LN4, 3LN5 - PubMed Abstract:
Polymorphic differences between human leukocyte antigen (HLA) molecules affect the specificity and conformation of their bound peptides and lead to differential selection of the T-cell repertoire. Mismatching during allogeneic transplantation can, therefore, lead to immunological reactions. We investigated the structure-function relationships of six members of the HLA-B*41 allelic group that differ by six polymorphic amino acids, including positions 80, 95, 97 and 114 within the antigen-binding cleft. Peptide-binding motifs for B*41:01, *41:02, *41:03, *41:04, *41:05 and *41:06 were determined by sequencing self-peptides from recombinant B*41 molecules by electrospray ionization tandem mass spectrometry. The crystal structures of HLA-B*41:03 bound to a natural 16-mer self-ligand (AEMYGSVTEHPSPSPL) and HLA-B*41:04 bound to a natural 11-mer self-ligand (HEEAVSVDRVL) were solved. Peptide analysis revealed that all B*41 alleles have an identical anchor motif at peptide position 2 (glutamic acid), but differ in their choice of C-terminal pΩ anchor (proline, valine, leucine). Additionally, B*41:04 displayed a greater preference for long peptides (>10 residues) when compared to the other B*41 allomorphs, while the longest peptide to be eluted from the allelic group (a 16mer) was obtained from B*41:03. The crystal structures of HLA-B*41:03 and HLA-B*41:04 revealed that both alleles interact in a highly conserved manner with the terminal regions of their respective ligands, while micropolymorphism-induced changes in the steric and electrostatic properties of the antigen-binding cleft account for differences in peptide repertoire and auxiliary anchoring. Differences in peptide repertoire, and peptide length specificity reflect the significant functional evolution of these closely related allotypes and signal their importance in allogeneic transplantation, especially B*41:03 and B*41:04, which accommodate longer peptides, creating structurally distinct peptide-HLA complexes.
Organizational Affiliation:
Institute for Transfusion Medicine, Hannover Medical School, Hannover, Germany.