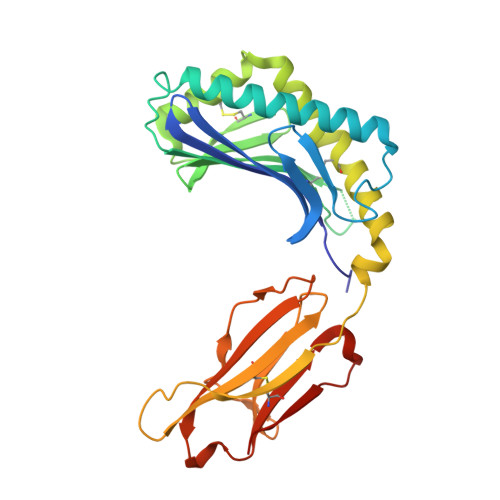
Crystal structure of bovine CD1b3 with endogenously bound ligands.
Girardi, E., Wang, J., Mac, T.T., Versluis, C., Bhowruth, V., Besra, G., Heck, A.J., Van Rhijn, I., Zajonc, D.M.(2010) J Immunol 185: 376-386
- PubMed: 20519644
- DOI: https://doi.org/10.4049/jimmunol.1000042
- Primary Citation of Related Structures:
3L9R - PubMed Abstract:
The CD1 family of Ag-presenting molecules is able to display lipids to T cells by binding them within a hydrophobic groove connected to the protein surface. In particular, the CD1b isotype is capable of binding ligands with greatly varying alkyl chain lengths through a complex network of interconnected hydrophobic pockets. Interestingly, mycobacterial lipids such as glucose monomycolate exclusively bind to CD1b. We determined the crystal structure of one of the three expressed bovine CD1b proteins, CD1b3, in complex with endogenous ligands, identified by mass spectrometry as a mixture of phosphatidylcholine and phosphatidylethanolamine, and analyzed the ability of the protein to bind glycolipids in vitro. The structure reveals a complex binding groove architecture, similar to the human ortholog but with consequential differences. Intriguingly, in bovine CD1b3 only the A', C' and F' pockets are present, whereas the T' pocket previously described in human CD1b is closed. This different pocket conformation could affect the ability of boCD1b3 to recognize lipids with long acyl chains such as glucose monomycolate. However, even in the absence of a T' tunnel, bovine CD1b3 is able to bind mycolates from Rhodococcus ruber in vitro.
Organizational Affiliation:
Division of Cell Biology, La Jolla Institute for Allergy and Immunology, La Jolla, CA, USA.