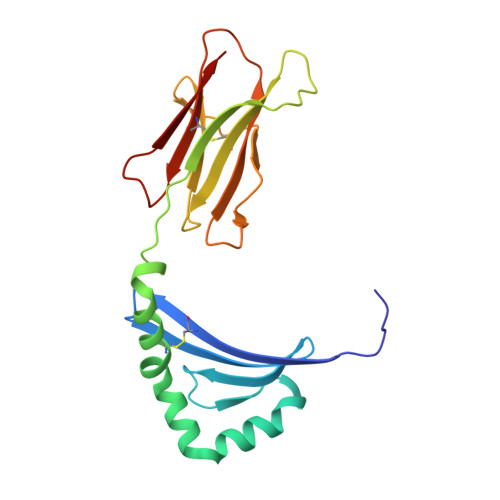
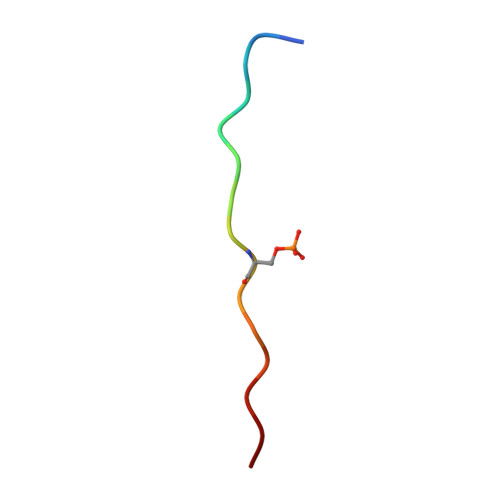
Structural Basis for the Presentation of Tumor-Associated MHC Class II-Restricted Phosphopeptides to CD4(+) T Cells.
Li, Y., Depontieu, F.R., Sidney, J., Salay, T.M., Engelhard, V.H., Hunt, D.F., Sette, A., Topalian, S.L., Mariuzza, R.A.(2010) J Mol Biol 399: 596-603
- PubMed: 20417641
- DOI: https://doi.org/10.1016/j.jmb.2010.04.037
- Primary Citation of Related Structures:
3L6F - PubMed Abstract:
Dysregulated protein phosphorylation is a hallmark of malignant transformation. Transformation can generate major histocompatibility complex (MHC)-bound phosphopeptides that are differentially displayed on tumor cells for specific recognition by T cells. To understand how phosphorylation alters the antigenic identity of self-peptides and how MHC class II molecules present phosphopeptides for CD4(+) T-cell recognition, we determined the crystal structure of a phosphopeptide derived from melanoma antigen recognized by T cells-1 (pMART-1), selectively expressed by human melanomas, in complex with HLA-DR1. The structure revealed that the phosphate moiety attached to the serine residue at position P5 of pMART-1 is available for direct interactions with T-cell receptor (TCR) and that the peptide N-terminus adopts an unusual conformation orienting it toward TCR. This structure, combined with measurements of peptide affinity for HLA-DR1 and of peptide-MHC recognition by pMART-1-specific T cells, suggests that TCR recognition is focused on the N-terminal portion of pMART-1. This recognition mode appears to be distinct from that of foreign antigen complexes but is remarkably reminiscent of the way autoreactive TCRs engage self- or altered self-peptides, consistent with the tolerogenic nature of tumor-host immune interactions.
Organizational Affiliation:
University of Maryland Institute for Bioscience and Biotechnology Research, W. M. Keck Laboratory for Structural Biology, Rockville, MD 20850, USA.