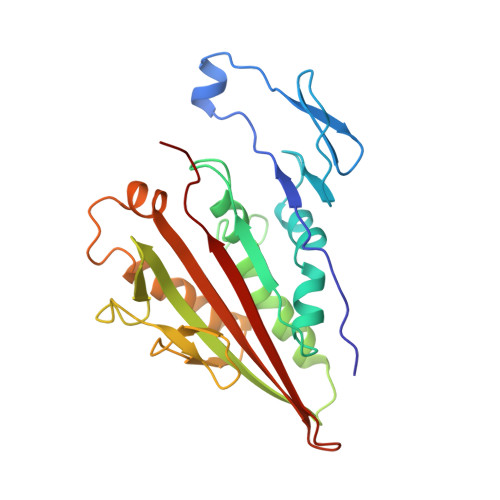
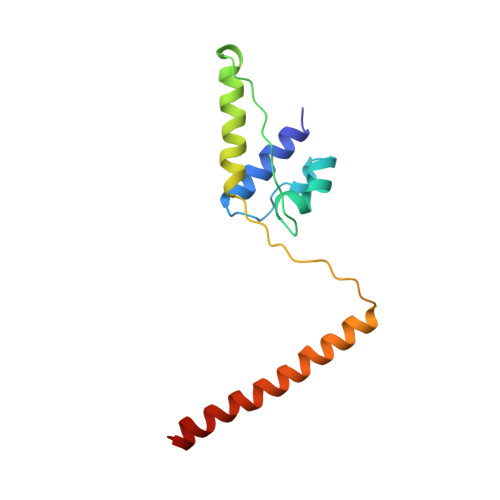
The crystal structure of dimeric kinesin and implications for microtubule-dependent motility.
Kozielski, F., Sack, S., Marx, A., Thormahlen, M., Schonbrunn, E., Biou, V., Thompson, A., Mandelkow, E.M., Mandelkow, E.(1997) Cell 91: 985-994
- PubMed: 9428521
- DOI: https://doi.org/10.1016/s0092-8674(00)80489-4
- Primary Citation of Related Structures:
3KIN - PubMed Abstract:
The dimeric form of the kinesin motor and neck domain from rat brain with bound ADP has been solved by X-ray crystallography. The two heads of the dimer are connected via a coiled-coil alpha-helical interaction of their necks. They are broadly similar to one another; differences are most apparent in the head-neck junction and in a moderate reorientation of the neck helices in order to adopt to the coiled-coil conformation. The heads show a rotational symmetry (approximately 120 degrees) about an axis close to that of the coiled-coil. This arrangement is unexpected since it is not compatible with the microtubule lattice. In this arrangement, the two heads of a kinesin dimer could not have equivalent interactions with microtubules.
Organizational Affiliation:
Max-Planck-Unit for Structural Molecular Biology, Hamburg, Germany.