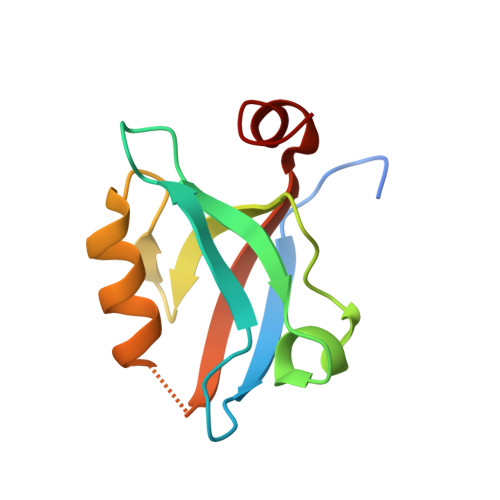
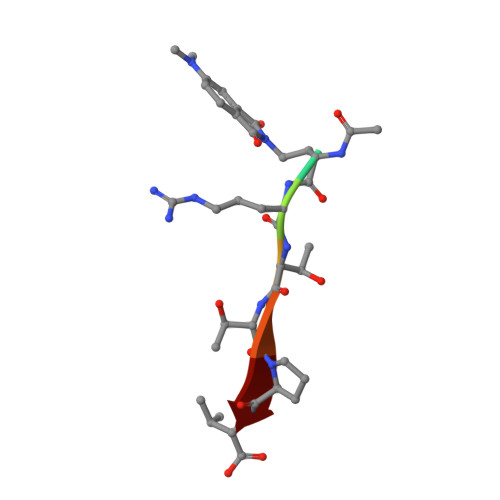
Caged mono- and divalent ligands for light-assisted disruption of PDZ domain-mediated interactions.
Sainlos, M., Iskenderian-Epps, W.S., Olivier, N.B., Choquet, D., Imperiali, B.(2013) J Am Chem Soc 135: 4580-4583
- PubMed: 23480637
- DOI: https://doi.org/10.1021/ja309870q
- Primary Citation of Related Structures:
3JXT - PubMed Abstract:
We report a general method for light-assisted control of interactions of PDZ domain binding motifs with their cognate domains by the incorporation of a photolabile caging group onto the essential C-terminal carboxylate binding determinant of the motif. The strategy was implemented and validated for both simple monovalent and biomimetic divalent ligands, which have recently been established as powerful tools for acute perturbation of native PDZ domain-dependent interactions in live cells.
Organizational Affiliation:
Departments of Chemistry and Biology, Massachusetts Institute of Technology, Cambridge, Massachusetts 02139, USA.