Crystallographic definition of the epitope promiscuity of the broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2F5: vaccine design implications.
Bryson, S., Julien, J.P., Hynes, R.C., Pai, E.F.(2009) J Virol 83: 11862-11875
- PubMed: 19740978
- DOI: https://doi.org/10.1128/JVI.01604-09
- Primary Citation of Related Structures:
1U8H, 1U8I, 1U8J, 1U8L, 1U8M, 1U8N, 1U8O, 1U8P, 1U8Q, 1U91, 1U92, 1U93, 1U95, 2F5A, 2F5B, 2PW1, 2PW2, 3IDG, 3IDI, 3IDJ, 3IDM, 3IDN - PubMed Abstract:
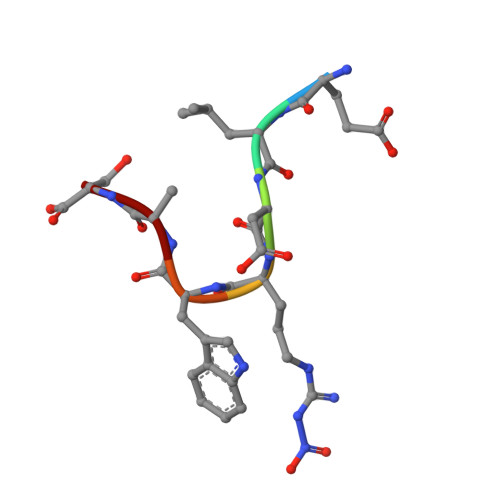
The quest to create a human immunodeficiency virus type 1 (HIV-1) vaccine capable of eliciting broadly neutralizing antibodies against Env has been challenging. Among other problems, one difficulty in creating a potent immunogen resides in the substantial overall sequence variability of the HIV envelope protein. The membrane-proximal region (MPER) of gp41 is a particularly conserved tryptophan-rich region spanning residues 659 to 683, which is recognized by three broadly neutralizing monoclonal antibodies (bnMAbs), 2F5, Z13, and 4E10. In this study, we first describe the variability of residues in the gp41 MPER and report on the invariant nature of 15 out of 25 amino acids comprising this region. Subsequently, we evaluate the ability of the bnMAb 2F5 to recognize 31 varying sequences of the gp41 MPER at a molecular level. In 19 cases, resulting crystal structures show the various MPER peptides bound to the 2F5 Fab'. A variety of amino acid substitutions outside the 664DKW666 core epitope are tolerated. However, changes at the 664DKW666 motif itself are restricted to those residues that preserve the aspartate's negative charge, the hydrophobic alkyl-pi stacking arrangement between the beta-turn lysine and tryptophan, and the positive charge of the former. We also characterize a possible molecular mechanism of 2F5 escape by sequence variability at position 667, which is often observed in HIV-1 clade C isolates. Based on our results, we propose a somewhat more flexible molecular model of epitope recognition by bnMAb 2F5, which could guide future attempts at designing small-molecule MPER-like vaccines capable of eliciting 2F5-like antibodies.
Organizational Affiliation:
Department of Biochemistry, University of Toronto, 1 King's College Circle, Toronto, Ontario, Canada M5S 1A8.