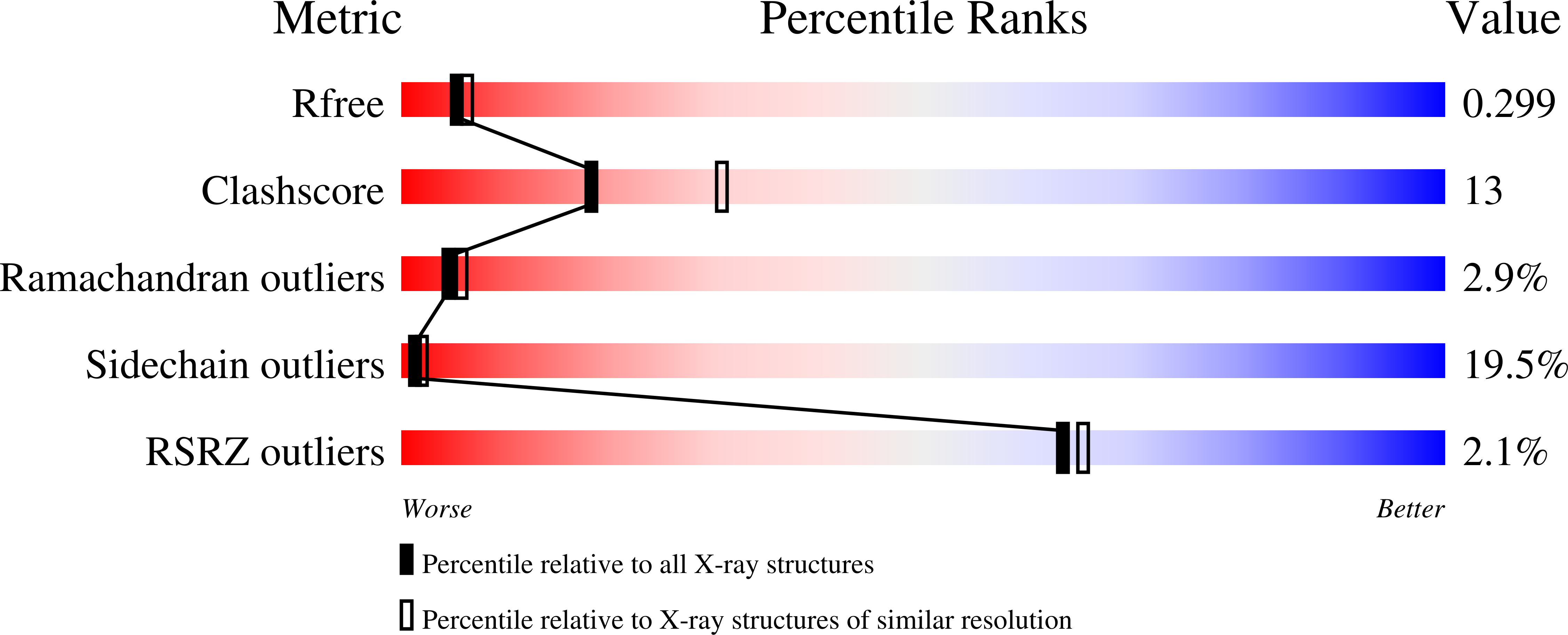
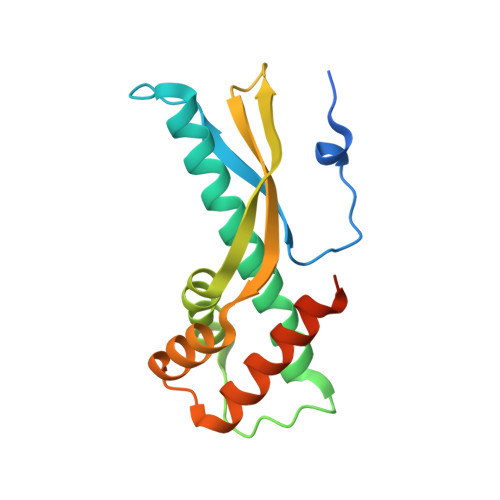
Structural basis for the Helicobacter pylori-carcinogenic TNF-alpha-inducing protein.
Tsuge, H., Tsurumura, T., Utsunomiya, H., Kise, D., Kuzuhara, T., Watanabe, T., Fujiki, H., Suganuma, M.(2009) Biochem Biophys Res Commun 388: 193-198
- PubMed: 19643085
- DOI: https://doi.org/10.1016/j.bbrc.2009.07.121
- Primary Citation of Related Structures:
3GUQ - PubMed Abstract:
Stomach cancer is strongly associated with infection by Helicobacter pylori. In 2005, we identified a new H. pylori gene encoding a TNF-alpha inducing protein (Tipalpha) that acts as a carcinogenic factor. Tipalpha is secreted from H. pylori as a homodimer whose subunits are linked by disulfide bonds. We also characterized a Tipalpha deletion mutant (del-Tipalpha) that lacks the N-terminal six amino acid residues (LQACTC), including two cysteines (C5 and C7) that form disulfide bonds, but nonetheless shows a weak ability to induce TNF-alpha expression. Here we report that del-Tipalpha has a novel elongated structure containing a 40-A-long alpha helix, and forms a heart-shaped homodimer via non-covalent bonds. Moreover, their circular dichroism spectra strongly suggest that the structures of the del-Tipalpha and Tipalpha homodimers are very similar. del-Tipalpha's unique mode of dimer formation provides important insight into protein-protein interactions and into the mechanism underlying the carcinogenicity of H. pylori infection.
Organizational Affiliation:
Institute for Health Sciences, Tokushima Bunri University, Yamashiro-Cho, Tokushima 770-8514, Japan. tsuge@tokushima.bunri-u.ac.jp