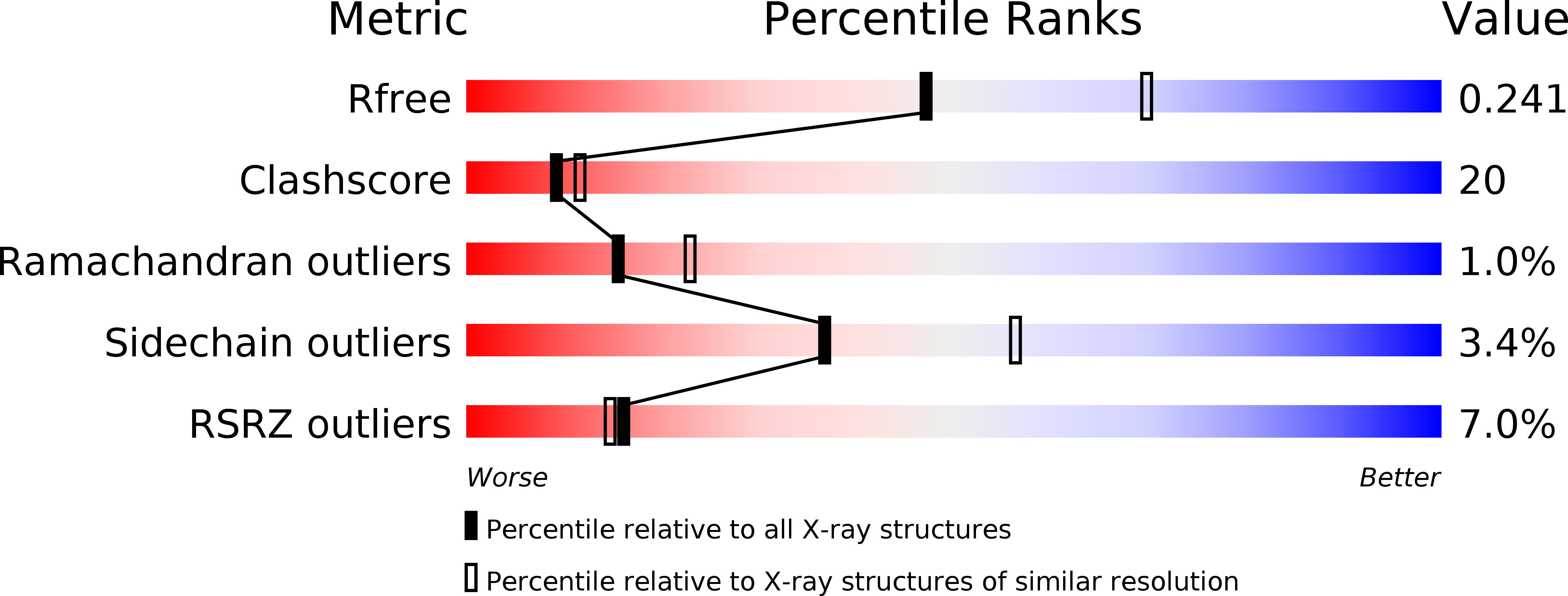
Crystal Structure of the TNF-alpha-Inducing Protein (Tipalpha) from Helicobacter pylori: Insights into Its DNA-Binding Activity.
Jang, J.Y., Yoon, H.J., Yoon, J.Y., Kim, H.S., Lee, S.J., Kim, K.H., Kim, D.J., Jang, S., Han, B.G., Lee, B.I., Suh, S.W.(2009) J Mol Biol
- PubMed: 19596016
- DOI: https://doi.org/10.1016/j.jmb.2009.07.010
- Primary Citation of Related Structures:
3GIO - PubMed Abstract:
Helicobacter pylori infection is one of the highest risk factors for gastroduodenal diseases including gastric cancer. Tumor necrosis factor-alpha (TNF-alpha) is one of the essential cytokines for tumor promotion, and thus, an H. pylori protein that induces TNF-alpha is believed to play a significant role in gastric cancer development in humans. The HP0596 gene product of H. pylori strain 26695 was identified as the TNF-alpha-inducing protein (Tipalpha). Tipalpha is secreted from H. pylori as dimers and enters the gastric cells. It was shown to have a DNA-binding activity. Here, we have determined the crystal structure of Tipalpha from H. pylori. Its monomer consists of two structural domains ("mixed domain" and "helical domain"). Tipalpha exists as a dimer in the crystal, and the dimeric structure represents a novel scaffold for DNA binding. A positively charged surface patch formed across the two monomers of the Tipalpha dimer by the loop between helices alpha1 and alpha2 may be important in DNA binding.
Organizational Affiliation:
Department of Chemistry, Seoul National University, Korea.