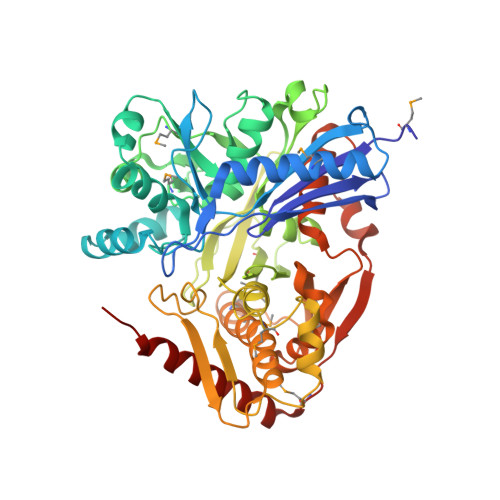
2.7 Angstrom Crystal Structure of Glycerol Kinase (glpK) from Staphylococcus aureus in Complex with ADP and Glycerol
Minasov, G., Brunzelle, J., Skarina, T., Onopriyenko, O., Peterson, S.N., Savchenko, A., Anderson, W.F., Center for Structural Genomics of Infectious Diseases (CSGID)To be published.