Antagonism of Secreted PCSK9 Increases Low Density Lipoprotein Receptor Expression in HepG2 Cells.
McNutt, M.C., Kwon, H.J., Chen, C., Chen, J.R., Horton, J.D., Lagace, T.A.(2009) J Biol Chem 284: 10561-10570
- PubMed: 19224862
- DOI: https://doi.org/10.1074/jbc.M808802200
- Primary Citation of Related Structures:
3GCW, 3GCX - PubMed Abstract:
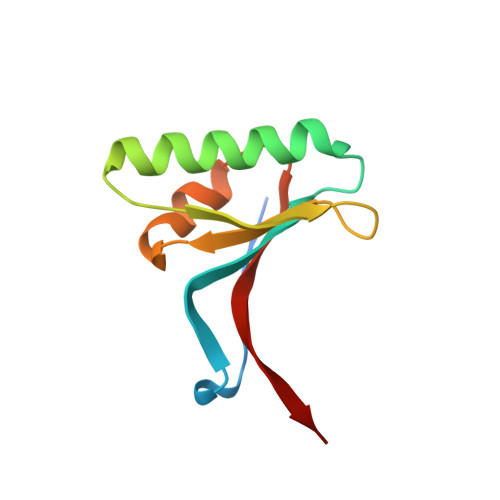
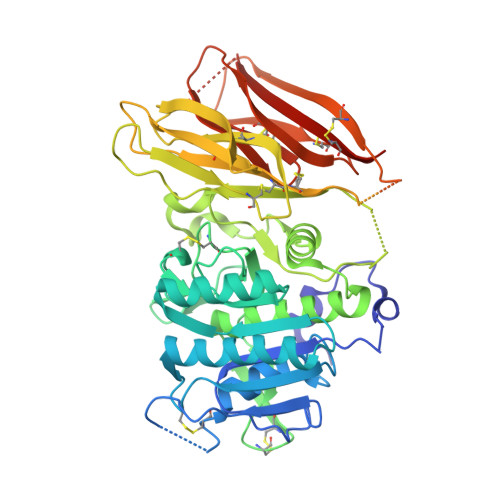
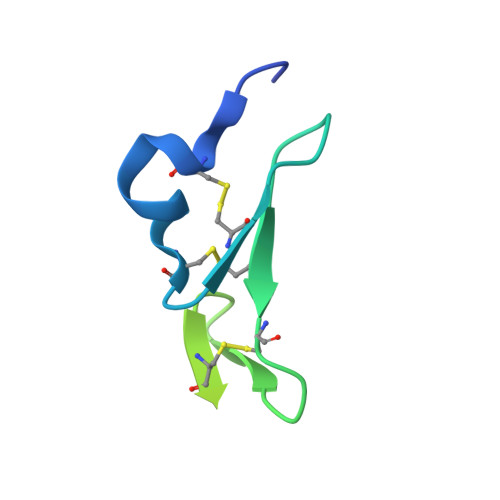
PCSK9 is a secreted protein that degrades low density lipoprotein receptors (LDLRs) in liver by binding to the epidermal growth factor-like repeat A (EGF-A) domain of the LDLR. It is not known whether PCSK9 causes degradation of LDLRs within the secretory pathway or following secretion and reuptake via endocytosis. Here we show that a mutation in the LDLR EGF-A domain associated with familial hypercholesterolemia, H306Y, results in increased sensitivity to exogenous PCSK9-mediated cellular degradation because of enhanced PCSK9 binding affinity. The crystal structure of the PCSK9-EGF-A(H306Y) complex shows that Tyr-306 forms a hydrogen bond with Asp-374 in PCSK9 at neutral pH, which strengthens the interaction with PCSK9. To block secreted PCSK9 activity, LDLR (H306Y) subfragments were added to the medium of HepG2 cells stably overexpressing wild-type PCSK9 or gain-of-function PCSK9 mutants associated with hypercholesterolemia (D374Y or S127R). These subfragments blocked secreted PCSK9 binding to cell surface LDLRs and resulted in the recovery of LDLR levels to those of control cells. We conclude that PCSK9 acts primarily as a secreted factor to cause LDLR degradation. These studies support the concept that pharmacological inhibition of the PCSK9-LDLR interaction extracellularly will increase hepatic LDLR expression and lower plasma low density lipoprotein levels.
Organizational Affiliation:
Departments of Molecular Genetics, Biochemistry, and Internal Medicine, University of Texas Southwestern Medical Center, Dallas, Texas 75390, USA.