Structural and Functional Analysis of a C3b-specific Antibody That Selectively Inhibits the Alternative Pathway of Complement
Katschke, K.J., Stawicki, S., Yin, J., Steffek, M., Xi, H., Sturgeon, L., Hass, P.E., Loyet, K.M., Deforge, L., Wu, Y., van Lookeren Campagne, M., Wiesmann, C.(2009) J Biol Chem 284: 10473-10479
- PubMed: 19196712
- DOI: https://doi.org/10.1074/jbc.M809106200
- Primary Citation of Related Structures:
3G6J - PubMed Abstract:
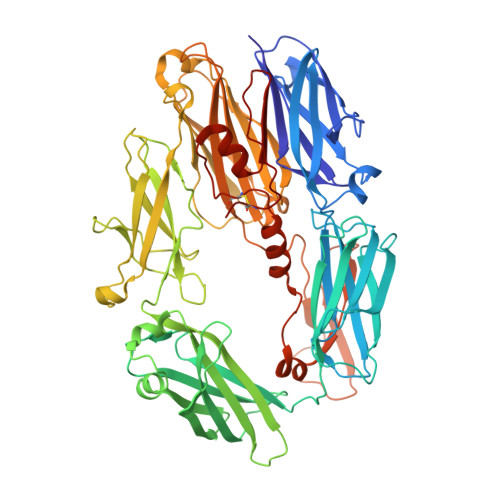
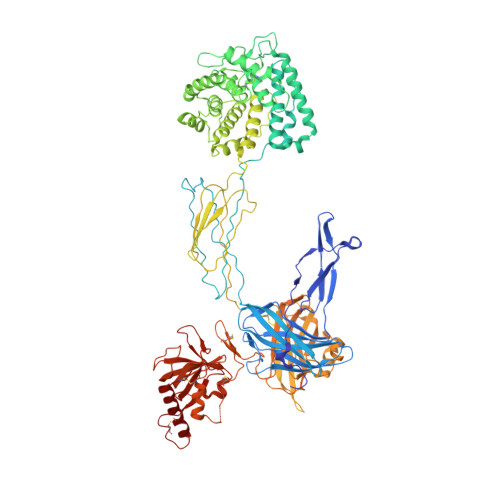
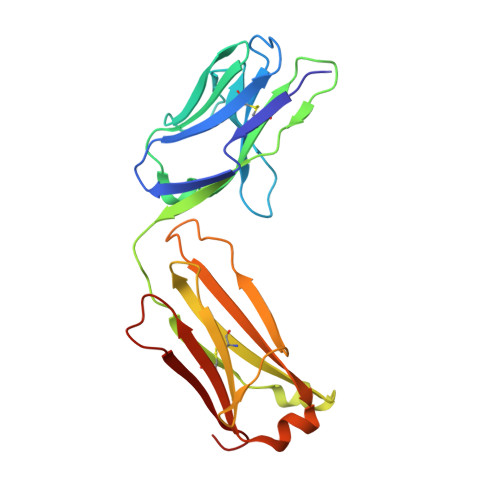
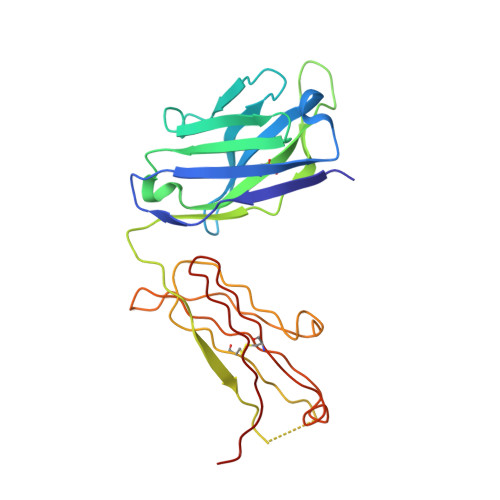
Amplification of the complement cascade through the alternative pathway can lead to excessive inflammation. Targeting C3b, a component central to the alternative pathway of complement, provides a powerful approach to inhibit complement-mediated immune responses and tissue injury. In the present study, phage display technology was employed to generate an antibody that selectively recognizes C3b but not the non-activated molecule C3. The crystal structure of C3b in complex with a Fab fragment of this antibody (S77) illustrates the structural basis for this selectivity. Cleavage of C3 to C3b results in a plethora of structural changes within C3, including the rearrangement of macroglobulin domain 6 enabling binding of S77 to the adjacent macroglobulin domain 7 domain. S77 blocks binding of factor B to C3b inhibiting the first step in the formation of the alternative pathway C3 convertase. In addition, S77 inhibits C5 binding to C3b. This results in significantly reduced formations of anaphylatoxins and membrane-attack complexes. This study for the first time demonstrates the structural basis for complement inhibition by a C3b-selective antibody and provides insights into the molecular mechanisms of alternative pathway complement activation.
Organizational Affiliation:
Departments of Immunology, Antibody Engineering, Protein Engineering, Assay & Automation Technology, and Protein Chemistry, Genentech Inc., 1 DNA Way, South San Francisco, California 94080, USA.