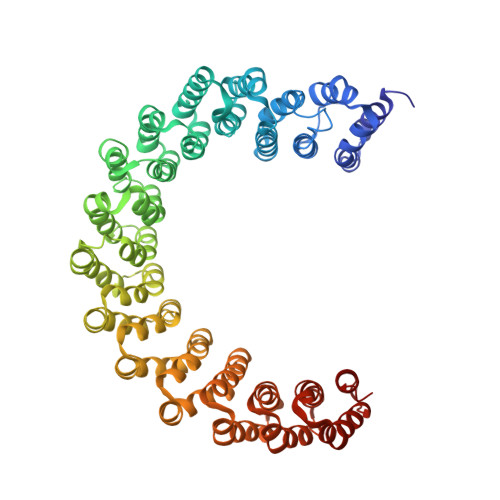Structure and function of the PP2A-shugoshin interaction
Xu, Z., Cetin, B., Anger, M., Cho, U.S., Helmhart, W., Nasmyth, K., Xu, W.(2009) Mol Cell 35: 426-441
- PubMed: 19716788
- DOI: https://doi.org/10.1016/j.molcel.2009.06.031
- Primary Citation of Related Structures:
3FGA - PubMed Abstract:
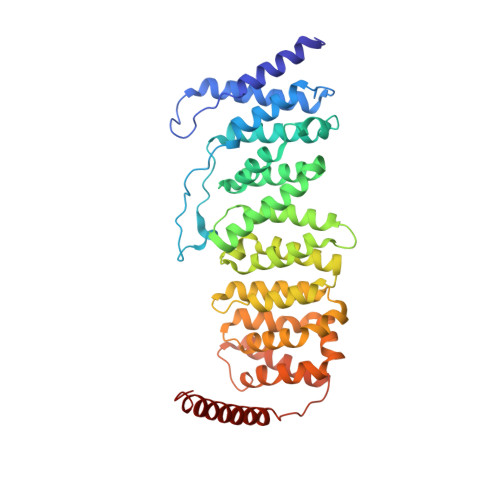
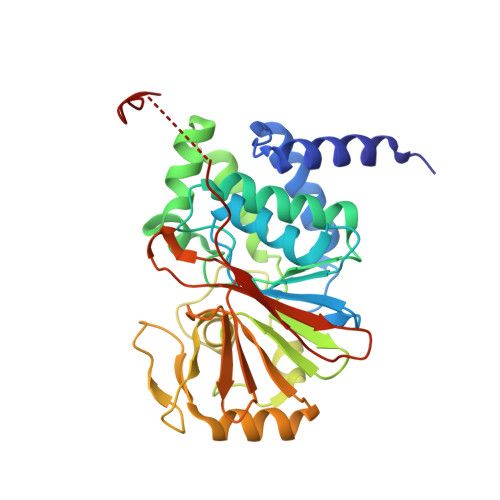
Accurate chromosome segregation during mitosis and meiosis depends on shugoshin proteins that prevent precocious dissociation of cohesin from centromeres. Shugoshins associate with PP2A, which is thought to dephosphorylate cohesin and thereby prevent cleavage by separase during meiosis I. A crystal structure of a complex between a fragment of human Sgo1 and an AB'C PP2A holoenzyme reveals that Sgo1 forms a homodimeric parallel coiled coil that docks simultaneously onto PP2A's C and B' subunits. Sgo1 homodimerization is a prerequisite for PP2A binding. While hSgo1 interacts only with the AB'C holoenzymes, its relative, Sgo2, interacts with all PP2A forms and may thus lead to dephosphorylation of distinct substrates. Mutant shugoshin proteins defective in the binding of PP2A cannot protect centromeric cohesin from separase during meiosis I or support the spindle assembly checkpoint in yeast. Finally, we provide evidence that PP2A's recruitment to chromosomes may be sufficient to protect cohesin from separase in mammalian oocytes.
Organizational Affiliation:
Department of Biological Structure, University of Washington, Seattle, WA 98195, USA.