Group B Streptococcus pullulanase crystal structures in the context of a novel strategy for vaccine development
Gourlay, L.J., Santi, I., Pezzicoli, A., Grandi, G., Soriani, M., Bolognesi, M.(2009) J Bacteriol 191: 3544-3552
- PubMed: 19329633
- DOI: https://doi.org/10.1128/JB.01755-08
- Primary Citation of Related Structures:
3FAW, 3FAX - PubMed Abstract:
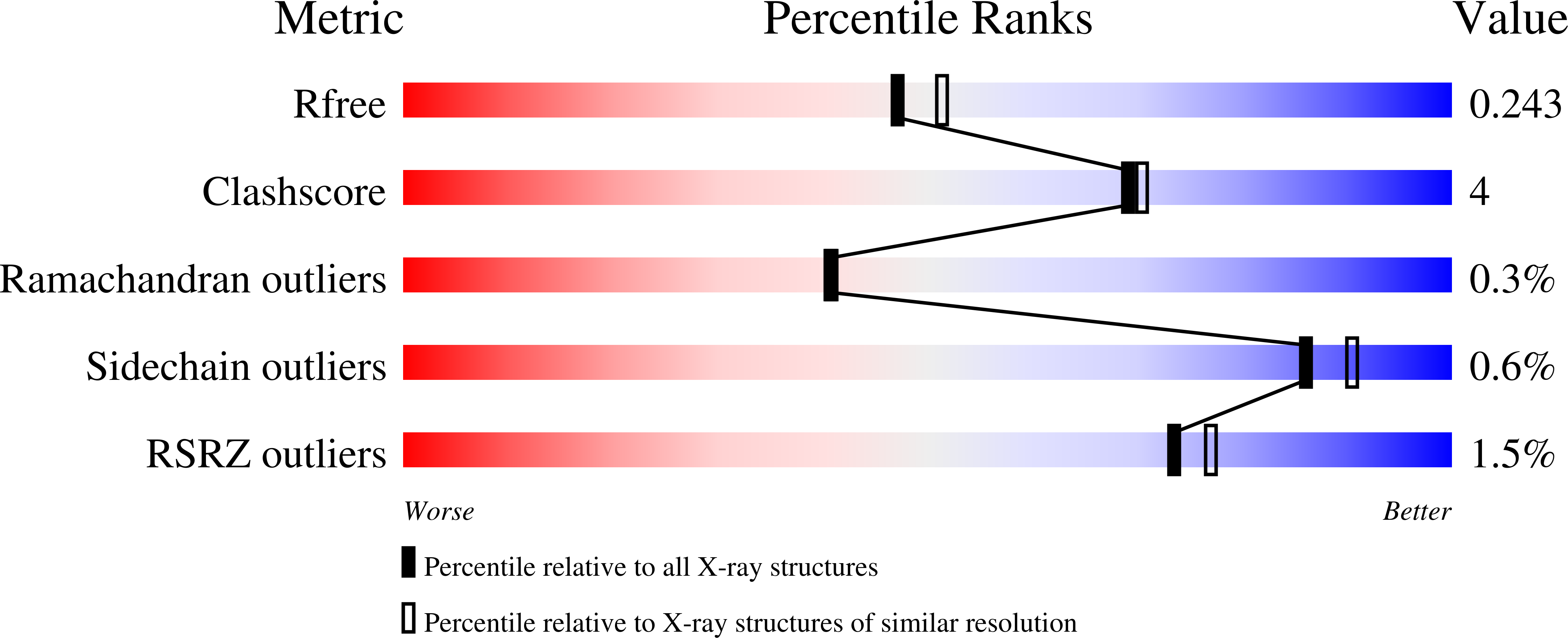
The group B streptococcus type I pullulanase (SAP) is a class 13 glycoside hydrolase that is anchored to the bacterial cell surface via a conserved C-terminal anchoring motif and involved in alpha-glucan degradation. Recent in vitro functional studies have shown that SAP is immunogenic in humans and that anti-SAP sera derived from immunized animals impair both group A and group B streptococcus pullulanase activities, suggesting that in vivo immunization with this antigen could prevent streptococcal colonization. To further investigate the putative role of SAP in bacterial pathogenesis, we carried out functional studies and found that recombinant SAP binds to human cervical epithelial cells. Furthermore, with a view of using SAP as a vaccine candidate, we present high-resolution crystal structure analyses of an N-terminally truncated form of SAP lacking the carbohydrate binding module but containing the catalytic domain and displaying glycosidase hydrolase activity, both in its apo form and in complex with maltotetraose, at resolutions of 2.1 and 2.4 A, respectively.
Organizational Affiliation:
Department of Biomolecular Sciences and Biotechnology, University of Milan, Via Celoria 26, I-20133 Milan, Italy.