Structural basis of the interaction between integrin alpha6beta4 and plectin at the hemidesmosomes
de Pereda, J.M., Lillo, M.P., Sonnenberg, A.(2009) EMBO J 28: 1180-1190
- PubMed: 19242489
- DOI: https://doi.org/10.1038/emboj.2009.48
- Primary Citation of Related Structures:
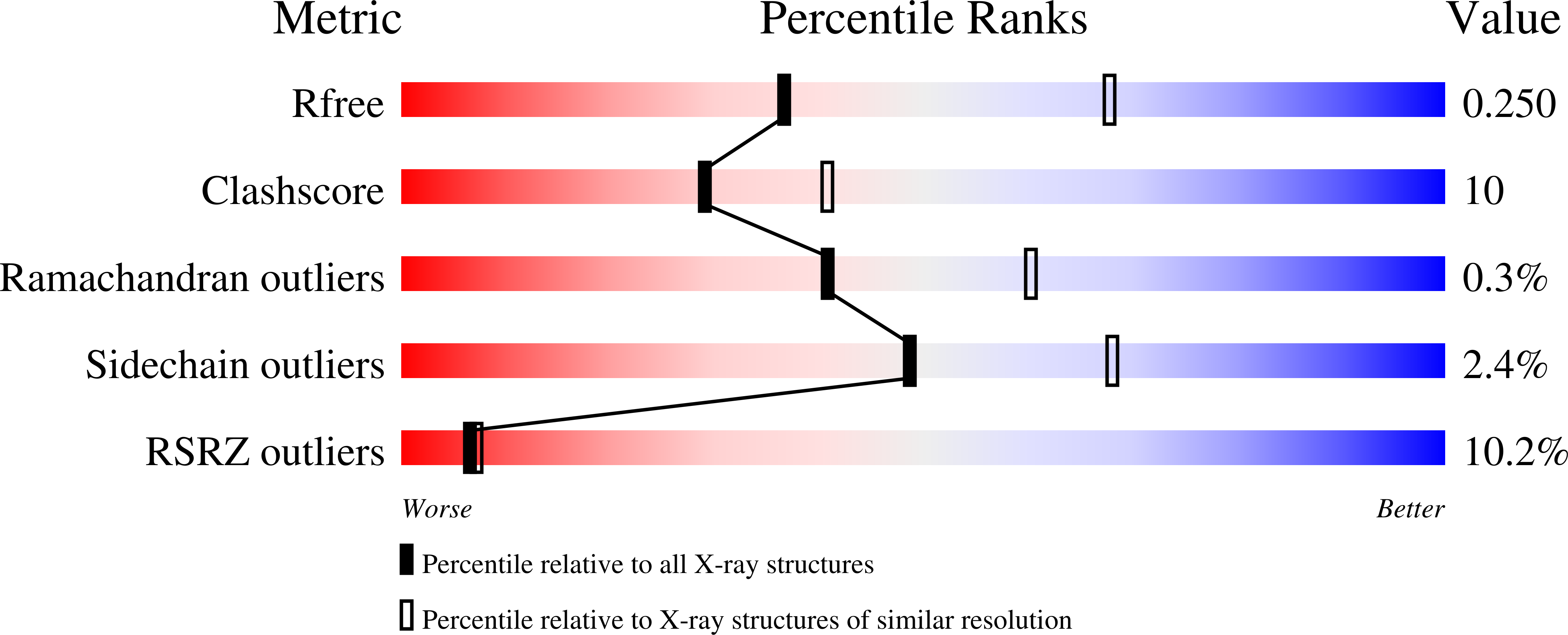
3F7P, 3F7Q, 3F7R - PubMed Abstract:
The interaction between the integrin alpha6beta4 and plectin is essential for the assembly and stability of hemidesmosomes, which are junctional adhesion complexes that anchor epithelial cells to the basement membrane. We describe the crystal structure at 2.75 A resolution of the primary alpha6beta4-plectin complex, formed by the first pair of fibronectin type III domains and the N-terminal region of the connecting segment of beta4 and the actin-binding domain of plectin. Two missense mutations in beta4 (R1225H and R1281W) linked to nonlethal forms of epidermolysis bullosa prevent essential intermolecular contacts. We also present two structures at 1.75 and 2.05 A resolution of the beta4 moiety in the absence of plectin, which reveal a major rearrangement of the connecting segment of beta4 on binding to plectin. This conformational switch is correlated with the way alpha6beta4 promotes stable adhesion or cell migration and suggests an allosteric control of the integrin.
Organizational Affiliation:
Department of Structural Biology, Instituto de Biología Molecular y Celular del Cáncer, Consejo Superior de Investigaciones Científicas-Universidad de Salamanca, Campus Unamuno, Salamanca, Spain. pereda@usal.es