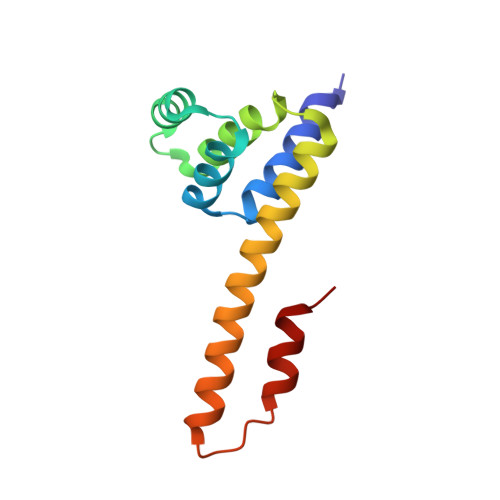
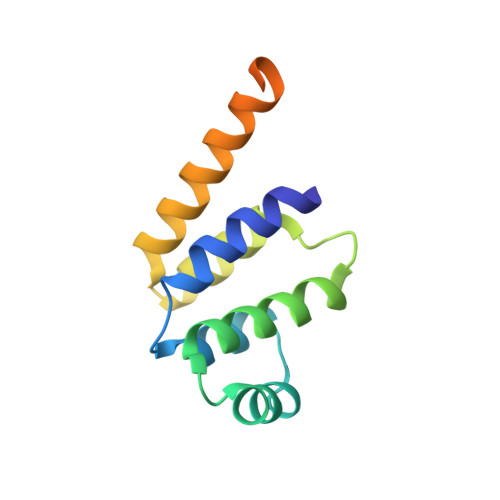
The Fas-FADD death domain complex structure unravels signalling by receptor clustering
Scott, F.L., Stec, B., Pop, C., Dobaczewska, M.K., Lee, J.J., Monosov, E., Robinson, H., Salvesen, G.S., Schwarzenbacher, R., Riedl, S.J.(2009) Nature 457: 1019-1022
- PubMed: 19118384
- DOI: https://doi.org/10.1038/nature07606
- Primary Citation of Related Structures:
3EZQ - PubMed Abstract:
The death inducing signalling complex (DISC) formed by Fas receptor, FADD (Fas-associated death domain protein) and caspase 8 is a pivotal trigger of apoptosis. The Fas-FADD DISC represents a receptor platform, which once assembled initiates the induction of programmed cell death. A highly oligomeric network of homotypic protein interactions comprised of the death domains of Fas and FADD is at the centre of DISC formation. Thus, characterizing the mechanistic basis for the Fas-FADD interaction is crucial for understanding DISC signalling but has remained unclear largely because of a lack of structural data. We have successfully formed and isolated the human Fas-FADD death domain complex and report the 2.7 A crystal structure. The complex shows a tetrameric arrangement of four FADD death domains bound to four Fas death domains. We show that an opening of the Fas death domain exposes the FADD binding site and simultaneously generates a Fas-Fas bridge. The result is a regulatory Fas-FADD complex bridge governed by weak protein-protein interactions revealing a model where the complex itself functions as a mechanistic switch. This switch prevents accidental DISC assembly, yet allows for highly processive DISC formation and clustering upon a sufficient stimulus. In addition to depicting a previously unknown mode of death domain interactions, these results further uncover a mechanism for receptor signalling solely by oligomerization and clustering events.
Organizational Affiliation:
Program in Apoptosis and Cell Death Research, The Burnham Institute for Medical Research, La Jolla, California 92037, USA.