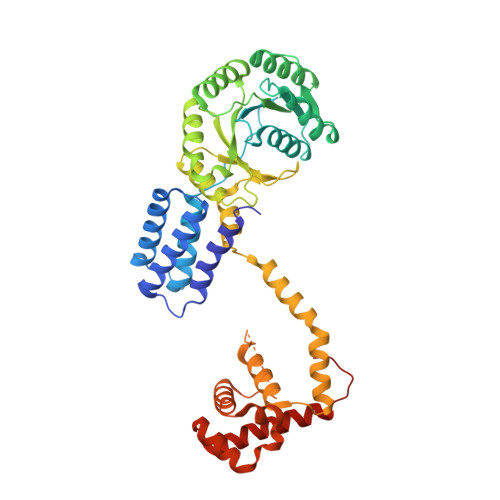
Structures of SRP54 and SRP19, the two proteins that organize the ribonucleic core of the signal recognition particle from Pyrococcus furiosus.
Egea, P.F., Napetschnig, J., Walter, P., Stroud, R.M.(2008) PLoS One 3: e3528-e3528
- PubMed: 18953414
- DOI: https://doi.org/10.1371/journal.pone.0003528
- Primary Citation of Related Structures:
3DLU, 3DLV, 3DM5 - PubMed Abstract:
In all organisms the Signal Recognition Particle (SRP), binds to signal sequences of proteins destined for secretion or membrane insertion as they emerge from translating ribosomes. In Archaea and Eucarya, the conserved ribonucleoproteic core is composed of two proteins, the accessory protein SRP19, the essential GTPase SRP54, and an evolutionarily conserved and essential SRP RNA. Through the GTP-dependent interaction between the SRP and its cognate receptor SR, ribosomes harboring nascent polypeptidic chains destined for secretion are dynamically transferred to the protein translocation apparatus at the membrane. We present here high-resolution X-ray structures of SRP54 and SRP19, the two RNA binding components forming the core of the signal recognition particle from the hyper-thermophilic archaeon Pyrococcus furiosus (Pfu). The 2.5 A resolution structure of free Pfu-SRP54 is the first showing the complete domain organization of a GDP bound full-length SRP54 subunit. In its ras-like GTPase domain, GDP is found tightly associated with the protein. The flexible linker that separates the GTPase core from the hydrophobic signal sequence binding M domain, adopts a purely alpha-helical structure and acts as an articulated arm allowing the M domain to explore multiple regions as it scans for signal peptides as they emerge from the ribosomal tunnel. This linker is structurally coupled to the GTPase catalytic site and likely to propagate conformational changes occurring in the M domain through the SRP RNA upon signal sequence binding. Two different 1.8 A resolution crystal structures of free Pfu-SRP19 reveal a compact, rigid and well-folded protein even in absence of its obligate SRP RNA partner. Comparison with other SRP19*SRP RNA structures suggests the rearrangement of a disordered loop upon binding with the RNA through a reciprocal induced-fit mechanism and supports the idea that SRP19 acts as a molecular scaffold and a chaperone, assisting the SRP RNA in adopting the conformation required for its optimal interaction with the essential subunit SRP54, and proper assembly of a functional SRP.
Organizational Affiliation:
Department of Biochemistry and Biophysics, University of California San Francisco, San Francisco, California, USA. pascal@msg.ucsf.edu