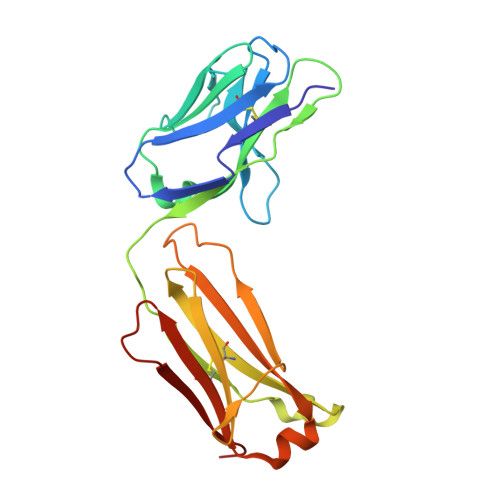
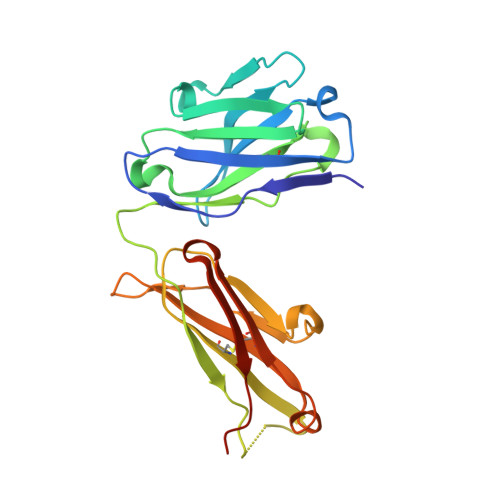
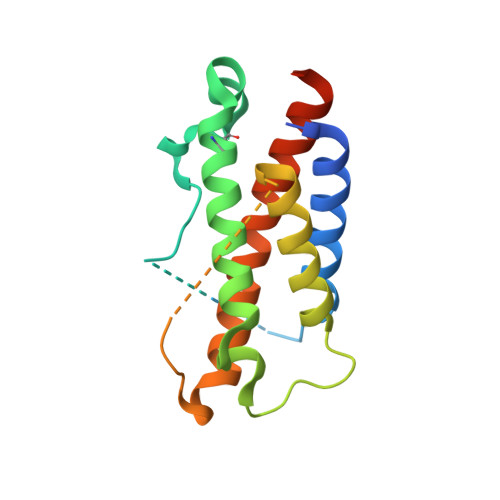
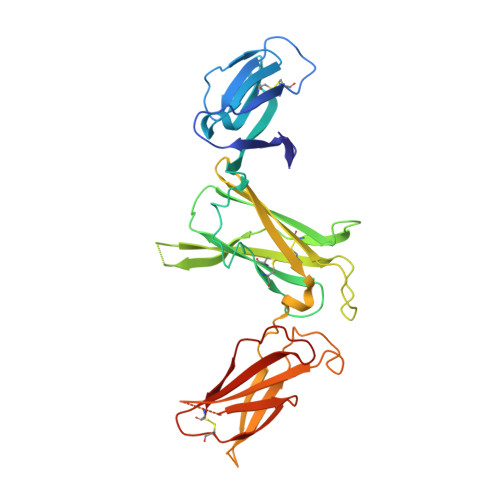
Crystal structures of the pro-inflammatory cytokine interleukin-23 and its complex with a high-affinity neutralizing antibody
Beyer, B.M., Ingram, R., Ramanathan, L., Reichert, P., Le, H.V., Madison, V., Orth, P.(2008) J Mol Biol 382: 942-955
- PubMed: 18708069
- DOI: https://doi.org/10.1016/j.jmb.2008.08.001
- Primary Citation of Related Structures:
3D85, 3D87 - PubMed Abstract:
Interleukin (IL)-23 is a pro-inflammatory cytokine playing a key role in the pathogenesis of several autoimmune and inflammatory diseases. We have determined the crystal structures of the heterodimeric p19-p40 IL-23 and its complex with the Fab (antigen-binding fragment) of a neutralizing antibody at 2.9 and 1.9 A, respectively. The IL-23 structure closely resembles that of IL-12. They share the common p40 subunit, and IL-23 p19 overlaps well with IL-12 p35. Along the hydrophilic heterodimeric interface, fewer charged residues are involved for IL-23 compared with IL-12. The binding site of the Fab is located exclusively on the p19 subunit, and comparison with published cytokine-receptor structures suggests that it overlaps with the IL-23 receptor binding site.
Organizational Affiliation:
Schering-Plough Research Institute, 2015 Galloping Hill Road, Kenilworth, NJ 07033, USA.