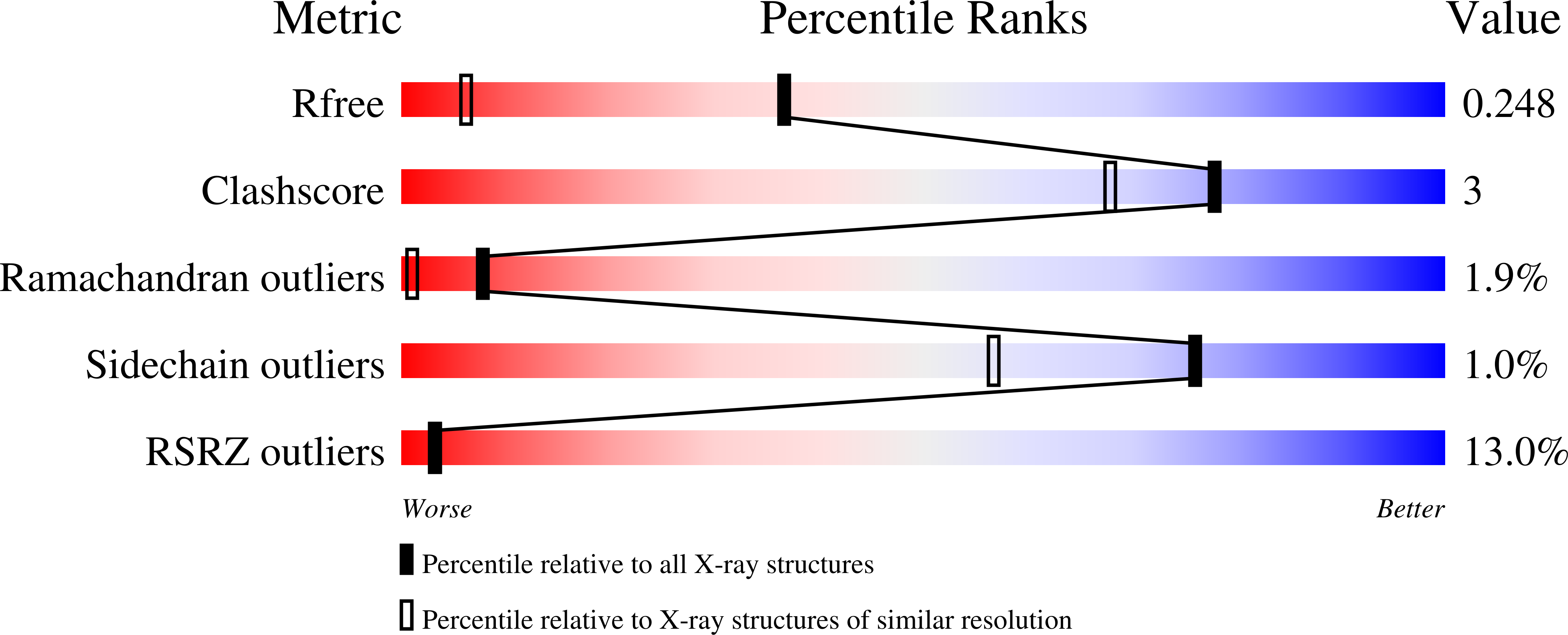
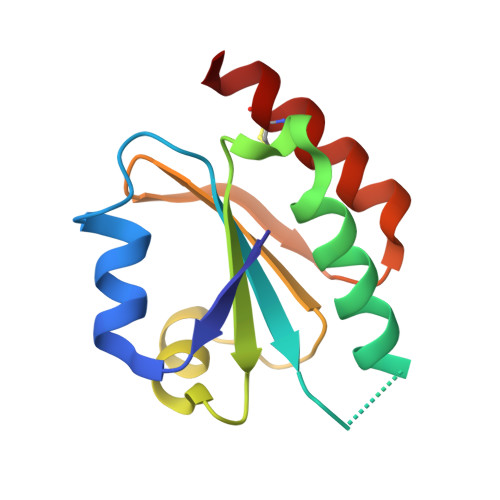
Structure of the thioredoxin-like domain of yeast glutaredoxin 3.
Gibson, L.M., Dingra, N.N., Outten, C.E., Lebioda, L.(2008) Acta Crystallogr D Biol Crystallogr 64: 927-932
- PubMed: 18703840
- DOI: https://doi.org/10.1107/S0907444908021641
- Primary Citation of Related Structures:
3D6I - PubMed Abstract:
Yeast glutaredoxin 3 (Grx3) is a cytosolic protein that regulates the activity of the iron-responsive transcriptional activator Aft1. This member of the monothiol glutaredoxin family contains a thioredoxin-like domain and a glutaredoxin-like domain, which both possess a monothiol active site. The crystal structure of the thioredoxin-like domain has been determined at 1.5 A resolution and represents the first published structure of this domain for the monothiol glutaredoxin family. The loop containing the signature motif WAxxC is partially disordered, indicating a greater degree of flexibility in this region compared with classical dithiol thioredoxins with a WCGPC active-site motif.
Organizational Affiliation:
Department of Chemistry and Biochemistry, University of South Carolina, Columbia, SC 29208, USA.