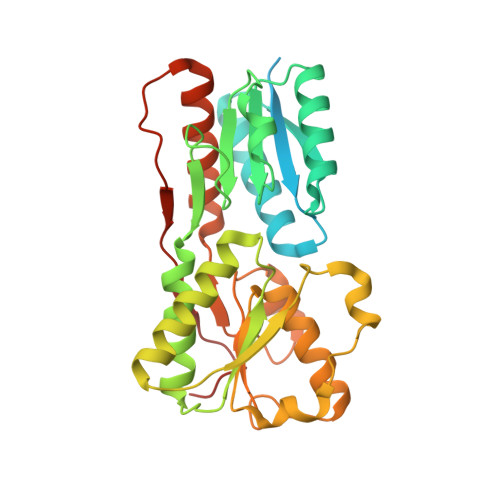
Crystal structure of periplasmic binding protein/LacI transcriptional regulator from Alkaliphilus metalliredigens QYMF complexed with L-xylulose.
Malashkevich, V.N., Toro, R., Wasserman, S.R., Meyer, A., Sauder, J.M., Burley, S.K., Almo, S.C.To be published.