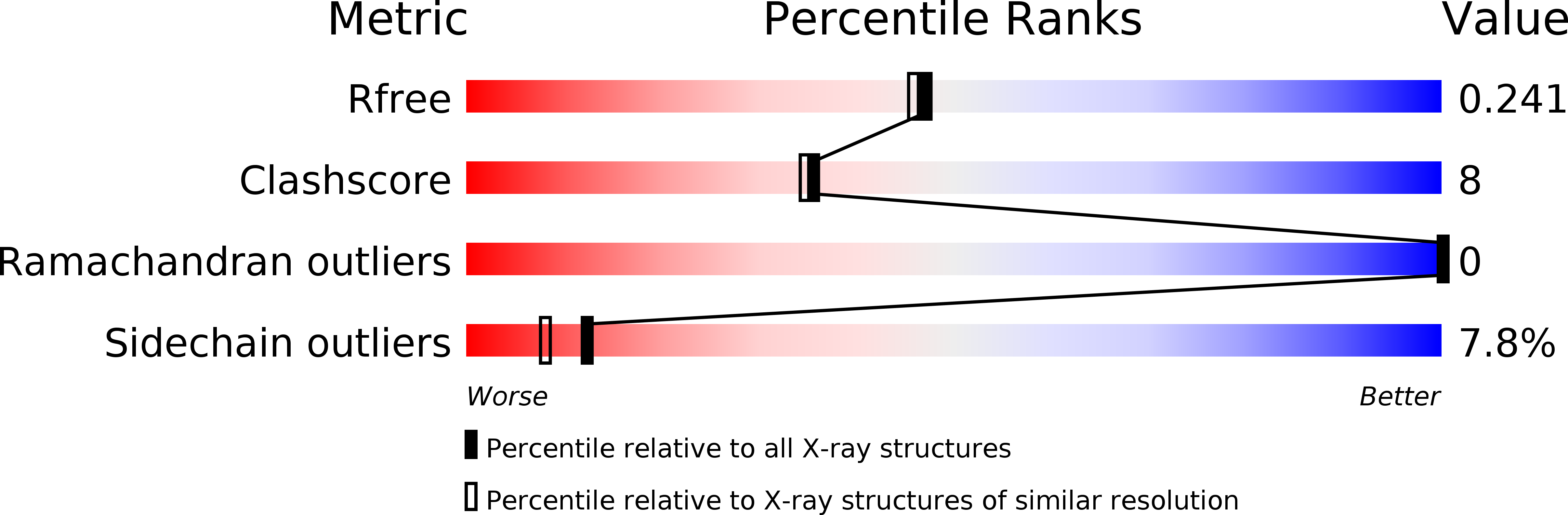The crystal structure of the heparin-binding reelin-N domain of f-spondin.
Tan, K., Duquette, M., Liu, J.H., Lawler, J., Wang, J.H.(2008) J Mol Biol 381: 1213-1223
- PubMed: 18602404
- DOI: https://doi.org/10.1016/j.jmb.2008.06.045
- Primary Citation of Related Structures:
3COO - PubMed Abstract:
The extracellular matrix protein F-spondin mediates axon guidance during neuronal development. Its N-terminal domain, termed the reelin-N domain, is conserved in F-spondins, reelins, and other extracellular matrix proteins. In this study, a recombinant human reelin-N domain has been expressed, purified, and shown to bind heparin. The crystal structure of the reelin-N domain resolved to 2.0 A reveals a variant immunoglobulin-like fold and potential heparin-binding sites. Substantial conformational variations even in secondary structure are observed between the two chemically identical reelin-N domains in one crystallographic asymmetric unit. The variations may result from extensive, highly specific interactions across the interface of the two reelin-N domains. The calculated values of buried surface area and the interface's shape complementarity are consistent with the formation of a weak dimer. The homophilic asymmetric dimer can potentially offer advantages in binding to ligands such as glycosaminoglycans, which may, in turn, bridge the two reelin-N domains and stabilize the dimer.
Organizational Affiliation:
Department of Medical Oncology, Dana-Farber Cancer Institute, Boston, MA 02115, USA. ktan@anl.gov