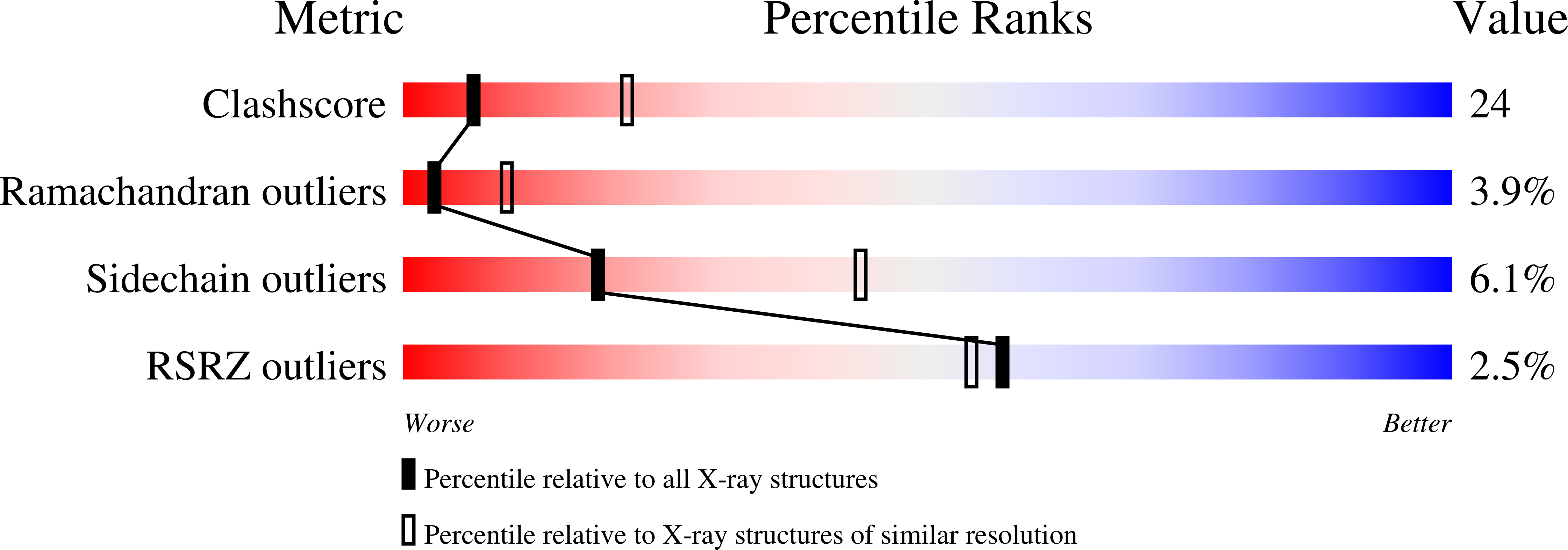Analyses of Mlc-IIBGlc interaction and a plausible molecular mechanism of Mlc inactivation by membrane sequestration
Nam, T.W., Jung, H.I., An, Y.J., Park, Y.H., Lee, S.H., Seok, Y.J., Cha, S.S.(2008) Proc Natl Acad Sci U S A 105: 3751-3756
- PubMed: 18319344
- DOI: https://doi.org/10.1073/pnas.0709295105
- Primary Citation of Related Structures:
3BP3, 3BP8 - PubMed Abstract:
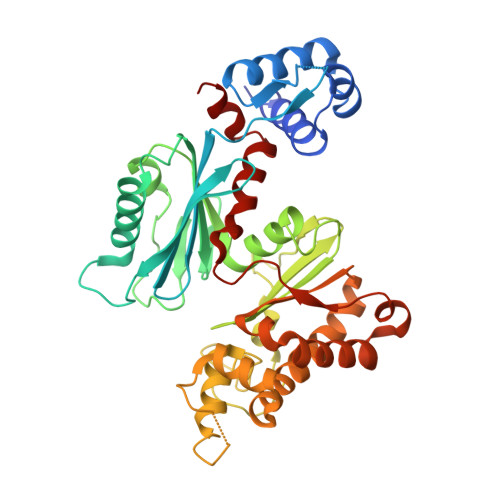
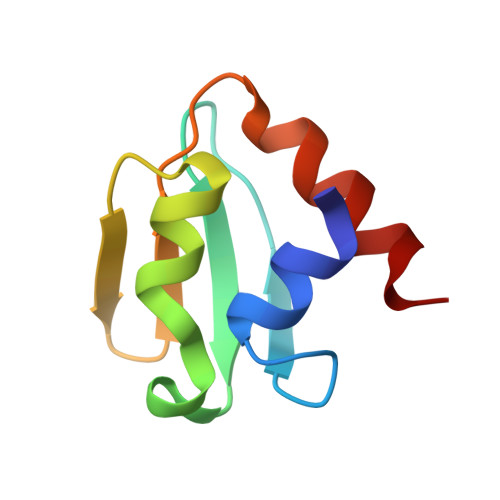
In Escherichia coli, glucose-dependent transcriptional induction of genes encoding a variety of sugar-metabolizing enzymes and transport systems is mediated by the phosphorylation state-dependent interaction of membrane-bound enzyme IICB(Glc) (EIICB(Glc)) with the global repressor Mlc. Here we report the crystal structure of a tetrameric Mlc in a complex with four molecules of enzyme IIB(Glc) (EIIB), the cytoplasmic domain of EIICB(Glc). Each monomer of Mlc has one bound EIIB molecule, indicating the 1:1 stoichiometry. The detailed view of the interface, along with the high-resolution structure of EIIB containing a sulfate ion at the phosphorylation site, suggests that the phosphorylation-induced steric hindrance and disturbance of polar intermolecular interactions impede complex formation. Furthermore, we reveal that Mlc possesses a built-in flexibility for the structural adaptation to its target DNA and that interaction of Mlc with EIIB fused only to dimeric proteins resulted in the loss of its DNA binding ability, suggesting that flexibility of the Mlc structure is indispensable for its DNA binding.
Organizational Affiliation:
Department of Biological Sciences and Institute of Microbiology, Seoul National University, Seoul 151-742, Korea.