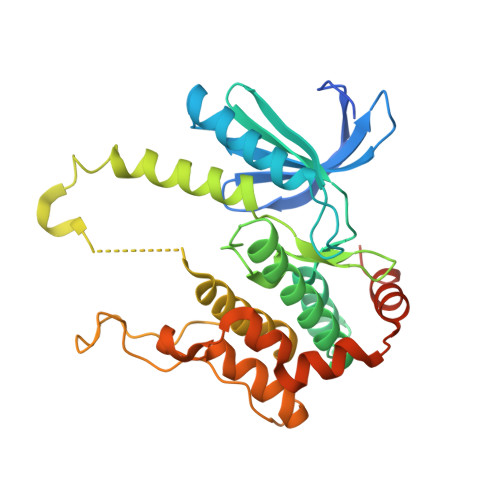
Crystal structures of MKK4 kinase domain reveal that substrate peptide binds to an allosteric site and induces an auto-inhibition state
Matsumoto, T., Kinoshita, T., Kirii, Y., Yokota, K., Hamada, K., Tada, T.(2010) Biochem Biophys Res Commun 400: 369-373
- PubMed: 20732303
- DOI: https://doi.org/10.1016/j.bbrc.2010.08.071
- Primary Citation of Related Structures:
3ALN, 3ALO - PubMed Abstract:
MKK4 activates both JNKs and p38s. We determined the crystal structures of human non-phosphorylated MKK4 kinase domain (npMKK4) complexed with AMP-PNP (npMKK4/AMP) and a ternary complex of npMKK4, AMP-PNP and p38α peptide (npMKK4/AMP/p38). These crystal structures revealed that the p38α peptide-bound npMKK4 at the allosteric site rather than at the putative substrate binding site and induced an auto-inhibition state. While the activation loop of the npMKK4/AMP complex was disordered, in the npMKK4/AMP/p38 complex it configured a long α-helix, which prevented substrate access to the active site and αC-helix movement to the active configuration of MKK4.
Organizational Affiliation:
PharmAxess, Inc., 3-9-12 Matsubara-cho, Akishima, Tokyo 196-8666, Japan.