Structural basis for the Rho- and phosphoinositide-dependent localization of the exocyst subunit Sec3
Yamashita, M., Kurokawa, K., Sato, Y., Yamagata, A., Mimura, H., Yoshikawa, A., Sato, K., Nakano, A., Fukai, S.(2010) Nat Struct Mol Biol 17: 180-186
- PubMed: 20062059
- DOI: https://doi.org/10.1038/nsmb.1722
- Primary Citation of Related Structures:
3A58 - PubMed Abstract:
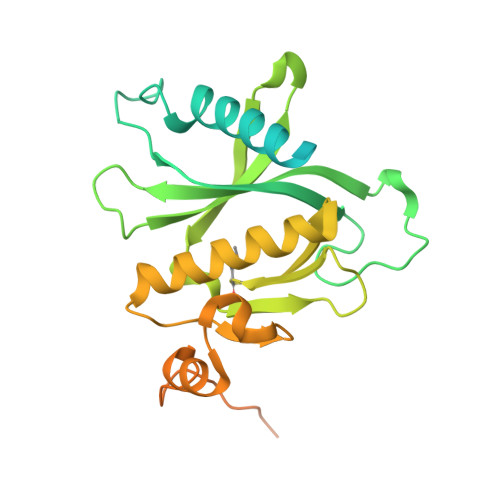
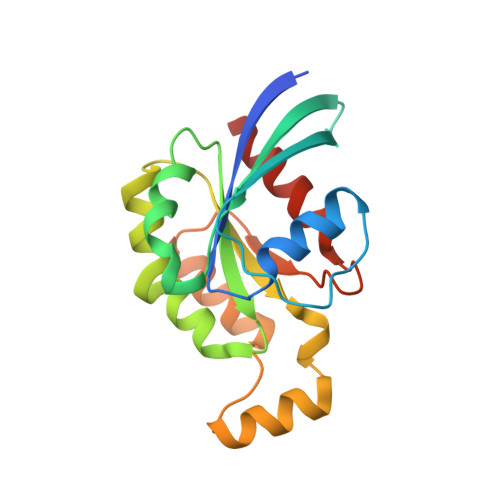
The exocyst complex is a hetero-octameric protein complex that functions during cell polarization by tethering the secretory vesicle to the target membrane. The yeast exocyst subunit Sec3 binds to phosphatidylinositol 4,5-bisphosphate (PtdIns(4,5)P(2)) and the small GTPases Rho1 and Cdc42 via its N-terminal domain (Sec3-N), and these interactions target Sec3 to the plasma membrane. Here we report the crystal structure of the Sec3-N in complex with Rho1 at 2.6-A resolution. Sec3-N adopts a pleckstrin homology (PH) fold, despite having no detectable sequence homology with other PH domains of known structure. Clusters of conserved basic residues constitute a positively charged cleft, which was identified as a binding site for PtdIns(4,5)P(2). Residues Phe77, Ile115 and Leu131 of Sec3 bind to an extended hydrophobic surface formed around switch regions I and II of Rho1. To our knowledge, these are the first structural insights into how an exocyst subunit might interact with both protein and phospholipid factors on the target membrane.
Organizational Affiliation:
Structural Biology Laboratory, Life Science Division, Synchrotron Radiation Research Organization and Institute of Molecular and Cellular Biosciences, The University of Tokyo, Tokyo, Japan.