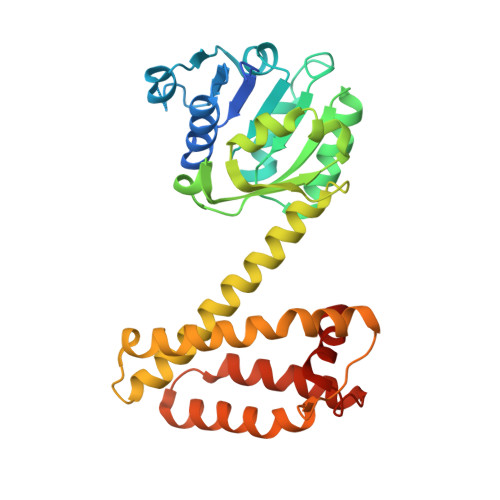
Structure and Activity of Nadph-Dependent Reductase Q1Eqe0 from Streptomyces Kanamyceticus, which Catalyses the R-Selective Reduction of an Imine Substrate.
Rodriguez-Mata, M., Frank, A., Wells, E., Leipold, F., Turner, N.J., Hart, S., Turkenburg, J.P., Grogan, G.(2013) Chembiochem 14: 1372
- PubMed: 23813853
- DOI: https://doi.org/10.1002/cbic.201300321
- Primary Citation of Related Structures:
3ZGY, 3ZHB - PubMed Abstract:
NADPH-dependent oxidoreductase Q1EQE0 from Streptomyces kanamyceticus catalyzes the asymmetric reduction of the prochiral monocyclic imine 2-methyl-1-pyrroline to the chiral amine (R)-2-methylpyrrolidine with >99% ee, and is thus of interest as a potential biocatalyst for the production of optically active amines. The structures of Q1EQE0 in native form, and in complex with the nicotinamide cofactor NADPH have been solved and refined to a resolution of 2.7 Å. Q1EQE0 functions as a dimer in which the monomer consists of an N-terminal Rossman-fold motif attached to a helical C-terminal domain through a helix of 28 amino acids. The dimer is formed through reciprocal domain sharing in which the C-terminal domains are swapped, with a substrate-binding cleft formed between the N-terminal subunit of monomer A and the C-terminal subunit of monomer B. The structure is related to those of known β-hydroxyacid dehydrogenases, except that the essential lysine, which serves as an acid/base in the (de)protonation of the nascent alcohol in those enzymes, is replaced by an aspartate residue, Asp187 in Q1EQE0. Mutation of Asp187 to either asparagine or alanine resulted in an inactive enzyme.
Organizational Affiliation:
York Structural Biology Laboratory, Department of Chemistry, University of York, Heslington, York, YO10 5DD, UK.