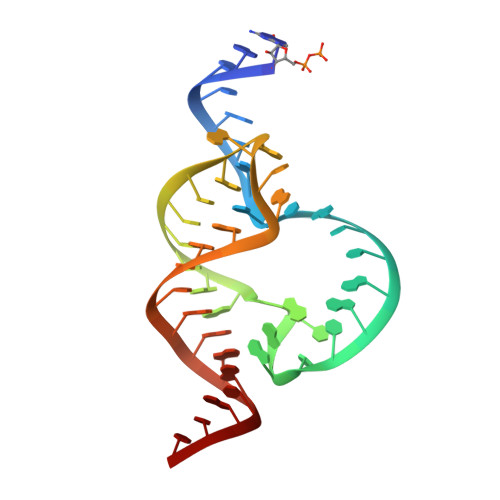
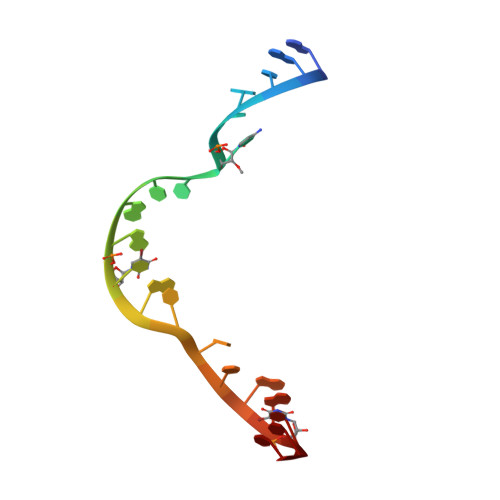
Tertiary contacts distant from the active site prime a ribozyme for catalysis.
Martick, M., Scott, W.G.(2006) Cell 126: 309-320
- PubMed: 16859740
- DOI: https://doi.org/10.1016/j.cell.2006.06.036
- Primary Citation of Related Structures:
3ZD5 - PubMed Abstract:
Minimal hammerhead ribozymes have been characterized extensively by static and time-resolved crystallography as well as numerous biochemical analyses, leading to mutually contradictory mechanistic explanations for catalysis. We present the 2.2 A resolution crystal structure of a full-length Schistosoma mansoni hammerhead ribozyme that permits us to explain the structural basis for its 1000-fold catalytic enhancement. The full-length hammerhead structure reveals how tertiary interactions occurring remotely from the active site prime this ribozyme for catalysis. G-12 and G-8 are positioned consistent with their previously suggested roles in acid-base catalysis, the nucleophile is aligned with a scissile phosphate positioned proximal to the A-9 phosphate, and previously unexplained roles of other conserved nucleotides become apparent within the context of a distinctly new fold that nonetheless accommodates the previous structural studies. These interactions permit us to explain the previously irreconcilable sets of experimental results in a unified, consistent, and unambiguous manner.
Organizational Affiliation:
Department of Molecular, Cellular and Developmental Biology, Robert L. Sinsheimer Laboratories, University of California, Santa Cruz, Santa Cruz, CA 95064, USA.