A Helical Rgd Motif Promoting Cell Adhesion: Crystal Structures of the Helicobacter Pylori Type Iv Secretion System Pilus Protein Cagl
Barden, S., Lange, S., Tegtmeyer, N., Conradi, J., Sewald, N., Backert, S., Niemann, H.H.(2013) Structure 21: 1931
- PubMed: 24076404
- DOI: https://doi.org/10.1016/j.str.2013.08.018
- Primary Citation of Related Structures:
3ZCI, 3ZCJ - PubMed Abstract:
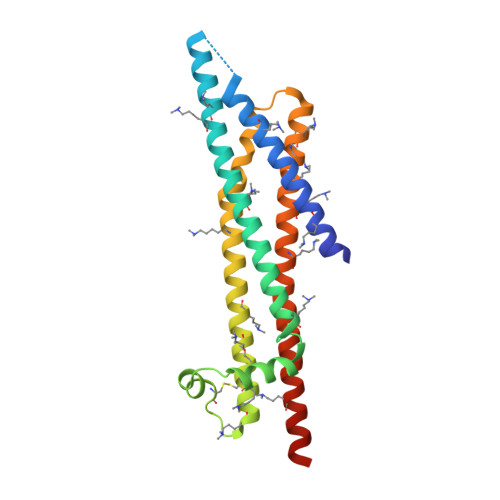
RGD tripeptide motifs frequently mediate ligand binding to integrins. The type IV secretion system (T4SS) protein CagL of the gastric pathogen Helicobacter pylori also contains an RGD motif. CagL decorates the T4SS pilus and may function as an adhesin for host cells. Whether CagL binds integrins via its RGD motif is under debate. Here, we present crystal structures of CagL revealing an elongated four-helix bundle that appears evolutionarily unrelated to the proposed VirB5 orthologs. The RGD motif is surface-exposed but located within a long α helix. This is unprecedented as previously characterized integrin-binding RGD motifs are located within extended or flexible loops. Yet, adhesion of gastric epithelial cells to CagL was strictly RGD-dependent. Comparison of seven crystallographically independent molecules reveals substantial structural flexibility. Intramolecular disulfide bonds engineered to reduce CagL flexibility resulted in more stable protein, but unable to support cell adhesion. CagL may thus partly unfold during receptor binding.
Organizational Affiliation:
Structural Biochemistry, Department of Chemistry, Bielefeld University, 33501 Bielefeld, Germany.