Toxicity Modulation, Resistance Enzyme Evasion, and A-Site X-ray Structure of Broad-Spectrum Antibacterial Neomycin Analogs
Maianti, J.P., Kanazawa, H., Dozzo, P., Matias, R.D., Feeney, L.A., Armstrong, E.S., Hildebrandt, D.J., Kane, T.R., Gliedt, M.J., Goldblum, A.A., Linsell, M.S., Aggen, J.B., Kondo, J., Hanessian, S.(2014) ACS Chem Biol 9: 2067-2073
- PubMed: 25019242
- DOI: https://doi.org/10.1021/cb5003416
- Primary Citation of Related Structures:
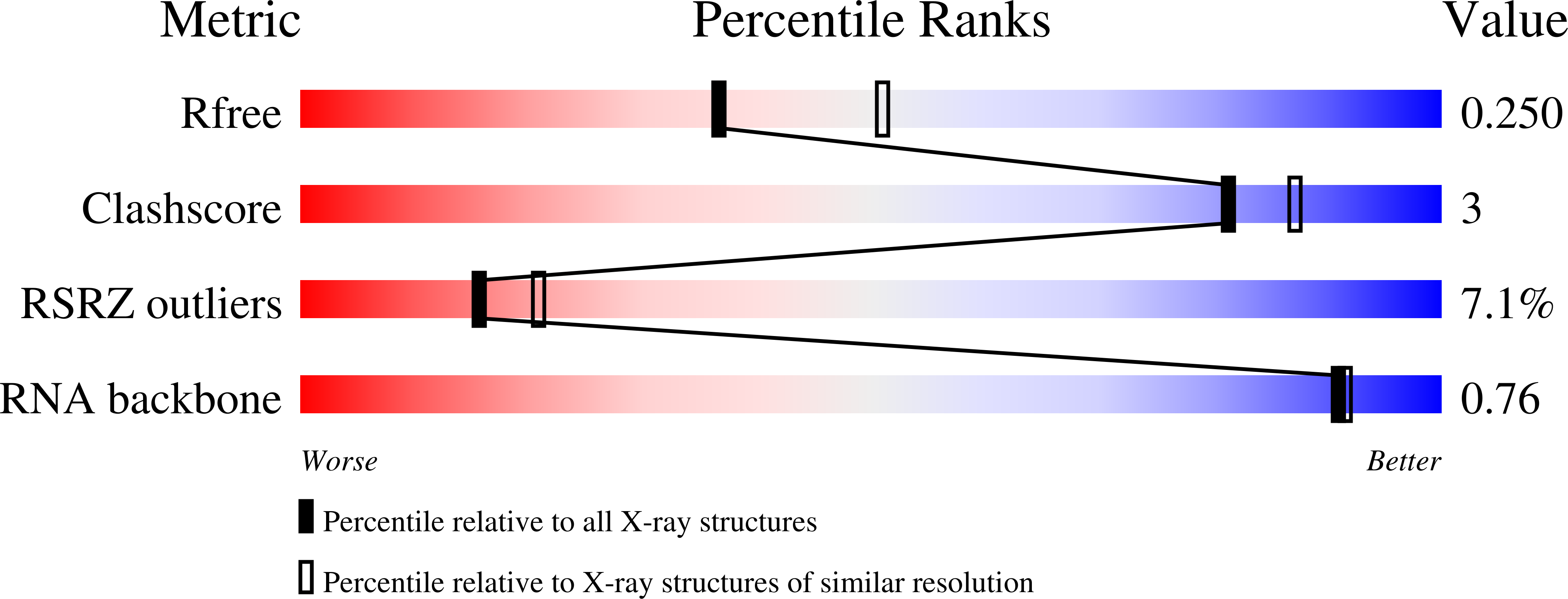
3WRU - PubMed Abstract:
Aminoglycoside antibiotics are pseudosaccharides decorated with ammonium groups that are critical for their potent broad-spectrum antibacterial activity. Despite over three decades of speculation whether or not modulation of pKa is a viable strategy to curtail aminoglycoside kidney toxicity, there is a lack of methods to systematically probe amine-RNA interactions and resultant cytotoxicity trends. This study reports the first series of potent aminoglycoside antibiotics harboring fluorinated N1-hydroxyaminobutyryl acyl (HABA) appendages for which fluorine-RNA contacts are revealed through an X-ray cocrystal structure within the RNA A-site. Cytotoxicity in kidney-derived cells was significantly reduced for the derivative featuring our novel β,β-difluoro-HABA group, which masks one net charge by lowering the pKa without compromising antibacterial potency. This novel side-chain assists in evasion of aminoglycoside-modifying enzymes, and it can be easily transferred to impart these properties onto any number of novel analogs.
Organizational Affiliation:
Department of Chemistry, Université de Montréal , CP 6128 Succ. Centre-Ville, Montréal, Québec H3C3J7, Canada.