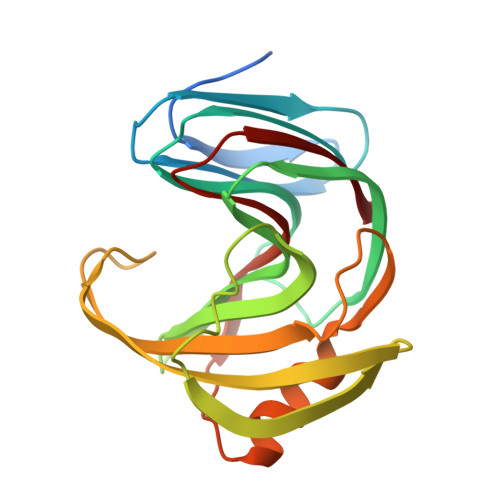
Crystal structure of Talaromyces cellulolyticus (formerly known as Acremonium cellulolyticus) GH family 11 xylanase
Kataoka, M., Akita, F., Maeno, Y., Inoue, B., Inoue, H., Ishikawa, K.(2014) Appl Biochem Biotechnol 174: 1599-1612
- PubMed: 25138599
- DOI: https://doi.org/10.1007/s12010-014-1130-9
- Primary Citation of Related Structures:
3WP3 - PubMed Abstract:
Talaromyces cellulolyticus (formerly known as Acremonium cellulolyticus) is one of the mesophilic fungi that can produce high levels of cellulose-related enzymes and are expected to be used for the degradation of polysaccharide biomass. In silico analysis of the genome sequence of T. cellulolyticus detected seven open reading frames (ORFs) showing homology to xylanases from glycoside hydrolase (GH) family 11. The gene encoding the GH11 xylanase C (TcXylC) with the highest activity was used for overproduction and purification of the recombinant enzyme, permitting solving of the crystal structure to a resolution of 1.98 Å. In the asymmetric unit, two kinds of the crystal structures of the xylanase were identified. The main structure of the protein showed a β-jelly roll structure. We hypothesize that one of the two structures represents the open form and the other shows the close form. The changing of the flexible region between the two structures is presumed to induce and accelerate the enzyme reaction. The specificity of xylanase toward the branched xylan is discussed in the context of this structural data and by comparison with the other published structures of xylanases.
Organizational Affiliation:
Biomass Refinery Research Center, National Institute of Advanced Industrial Science and Technology (AIST), 3-11-32, Kagamiyama, Higashihiroshima, Hiroshima, 739-0046, Japan.