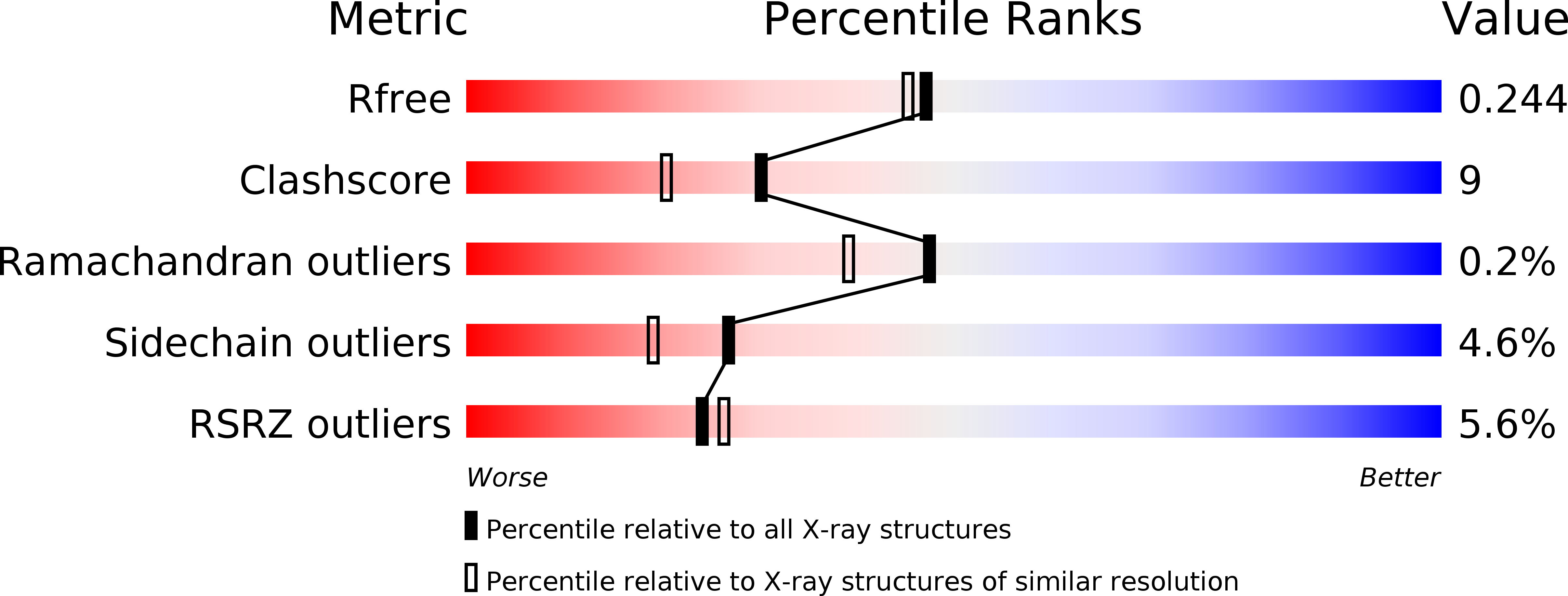
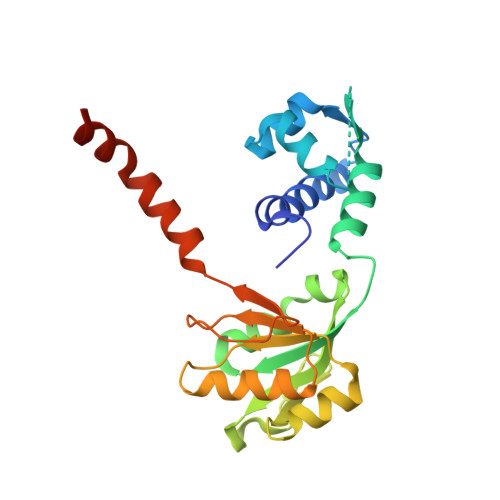
Distinct structural features of Rex-family repressors to sense redox levels in anaerobes and aerobes.
Zheng, Y., Ko, T.-P., Sun, H., Huang, C.-H., Pei, J., Qiu, R., Wang, A.H.-J., Wiegel, J., Shao, W., Guo, R.-T.(2014) J Struct Biol 188: 195-204
- PubMed: 25463021
- DOI: https://doi.org/10.1016/j.jsb.2014.11.001
- Primary Citation of Related Structures:
3WG9, 3WGH, 3WGI - PubMed Abstract:
The Rex-family repressors sense redox levels by alternative binding to NADH or NAD(+). Unlike other Rex proteins that regulate aerobic respiration, RSP controls ethanol fermentation in the obligate anaerobe Thermoanaerobacter ethanolicus JW200(T). It is also found in other anaerobic microorganisms. Here we present the crystal structures of apo-RSP, RSP/NADH and RSP/NAD(+)/DNA, which are the first structures of Rex-family members from an obligate anaerobe. RSP functions as a homodimer. It assumes an open conformation when bound to the operator DNA and a closed conformation when not DNA-bound. The DNA binds to the N-terminal winged-helix domain and the dinucleotide, either reduced or oxidized, binds to the C-terminal Rossmann-fold domain. The two distinct orientations of nicotinamide ring, anti in NADH and syn in NAD(+), give rise to two sets of protein-ligand interactions. Consequently, NADH binding makes RSP into a closed conformation, which does not bind to DNA. Both the conserved residues and the DNA specificity of RSP show a number of variations from those of the aerobic Rex, reflecting different structural bases for redox-sensing by the anaerobic and aerobic Rex-family members.
Organizational Affiliation:
Industrial Enzymes National Engineering Laboratory, Tianjin Institute of Industrial Biotechnology, Chinese Academy of Sciences, Tianjin 300308, China.