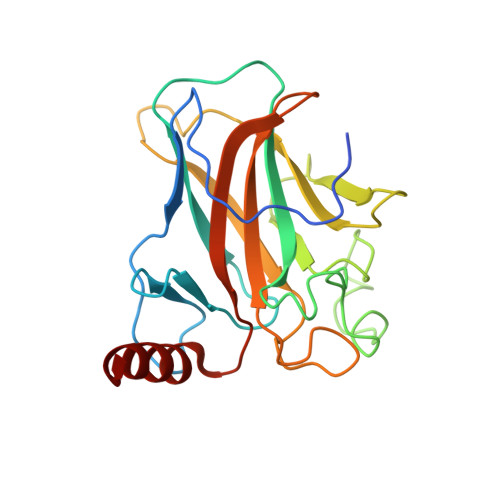
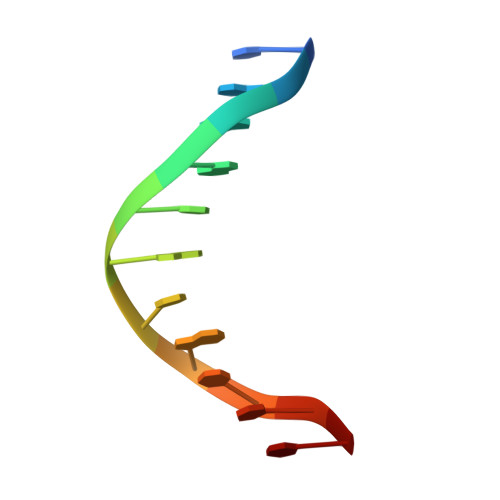
Structure of p73 DNA-binding domain tetramer modulates p73 transactivation.
Ethayathulla, A.S., Tse, P.W., Monti, P., Nguyen, S., Inga, A., Fronza, G., Viadiu, H.(2012) Proc Natl Acad Sci U S A 109: 6066-6071
- PubMed: 22474346
- DOI: https://doi.org/10.1073/pnas.1115463109
- Primary Citation of Related Structures:
3VD0, 3VD1, 3VD2 - PubMed Abstract:
The transcription factor p73 triggers developmental pathways and overlaps stress-induced p53 transcriptional pathways. How p53-family response elements determine and regulate transcriptional specificity remains an unsolved problem. In this work, we have determined the first crystal structures of p73 DNA-binding domain tetramer bound to response elements with spacers of different length. The structure and function of the adaptable tetramer are determined by the distance between two half-sites. The structures with zero and one base-pair spacers show compact p73 DNA-binding domain tetramers with large tetramerization interfaces; a two base-pair spacer results in DNA unwinding and a smaller tetramerization interface, whereas a four base-pair spacer hinders tetramerization. Functionally, p73 is more sensitive to spacer length than p53, with one base-pair spacer reducing 90% of transactivation activity and longer spacers reducing transactivation to basal levels. Our results establish the quaternary structure of the p73 DNA-binding domain required as a scaffold to promote transactivation.
Organizational Affiliation:
Laboratory of Structural Biochemistry, Department of Chemistry and Biochemistry, University of California at San Diego, 9500 Gilman Drive 0378, La Jolla, CA 92093, USA.