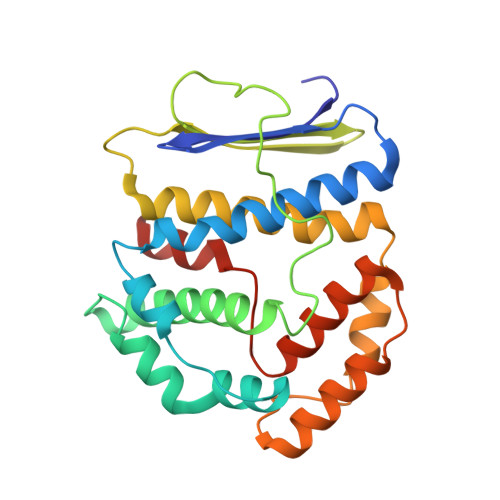
Crystal structure of Pyrococcus furiosus PF2050, a member of the DUF2666 protein family
Han, B.G., Jeong, K.C., Cho, J.W., Jeong, B.C., Song, H.K., Lee, J.Y., Shin, D.H., Lee, S., Lee, B.I.(2012) FEBS Lett 586: 1384-1388
- PubMed: 22616997
- DOI: https://doi.org/10.1016/j.febslet.2012.04.004
- Primary Citation of Related Structures:
3V68 - PubMed Abstract:
Pyrococcus furiosus PF2050 is an uncharacterized putative protein that contains two DUF2666 domains. Functional and structural studies of PF2050 have not previously been performed. In this study, we determined the crystal structure of PF2050. The structure of PF2050 showed that the two DUF2666 domains interact tightly, forming a globular structure. Each DUF2666 domain comprises an antiparallel β-sheet and an α-helical bundle. One side of the PF2050 structure has a positively charged basic cleft, which may have a DNA-binding function. Furthermore, we confirmed that PF2050 interacts with circular and linear dsDNA.
Organizational Affiliation:
Biomolecular Function Research Branch, Division of Convergence Technology, Research Institute, National Cancer Center, Goyang, Gyeonggi, Republic of Korea.