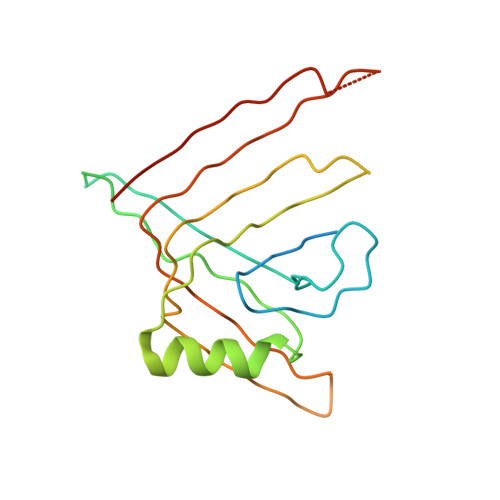
Crystal structure of a type VI secretion system effector from Yersinia pestis
Filippova, E.V., Halavaty, A., Minasov, G., Shuvalova, L., Dubrovska, I., Winsor, J., Papazisi, L., Anderson, W.F., Center for Structural Genomics of Infectious Diseases (CSGID)To be published.