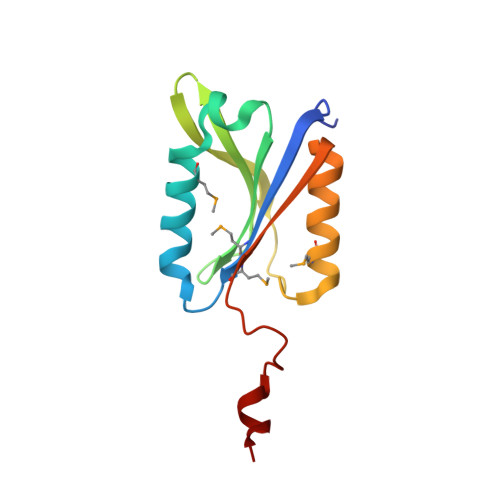
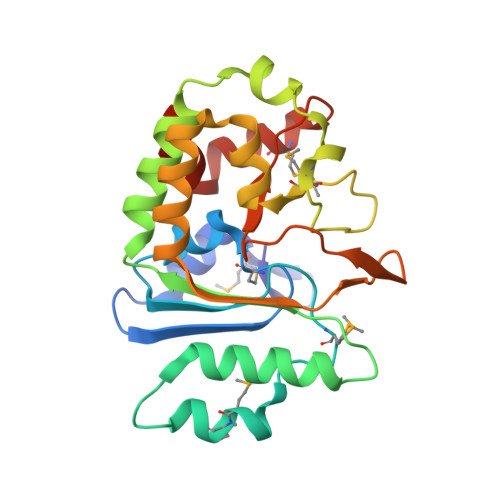
Crystal Structure and Activity Studies of the C11 Cysteine Peptidase from Parabacteroides merdae in the Human Gut Microbiome.
McLuskey, K., Grewal, J.S., Das, D., Godzik, A., Lesley, S.A., Deacon, A.M., Coombs, G.H., Elsliger, M.A., Wilson, I.A., Mottram, J.C.(2016) J Biol Chem 291: 9482-9491
- PubMed: 26940874
- DOI: https://doi.org/10.1074/jbc.M115.706143
- Primary Citation of Related Structures:
3UWS - PubMed Abstract:
Clan CD cysteine peptidases, a structurally related group of peptidases that include mammalian caspases, exhibit a wide range of important functions, along with a variety of specificities and activation mechanisms. However, for the clostripain family (denoted C11), little is currently known. Here, we describe the first crystal structure of a C11 protein from the human gut bacterium, Parabacteroides merdae (PmC11), determined to 1.7-Å resolution. PmC11 is a monomeric cysteine peptidase that comprises an extended caspase-like α/β/α sandwich and an unusual C-terminal domain. It shares core structural elements with clan CD cysteine peptidases but otherwise structurally differs from the other families in the clan. These studies also revealed a well ordered break in the polypeptide chain at Lys(147), resulting in a large conformational rearrangement close to the active site. Biochemical and kinetic analysis revealed Lys(147) to be an intramolecular processing site at which cleavage is required for full activation of the enzyme, suggesting an autoinhibitory mechanism for self-preservation. PmC11 has an acidic binding pocket and a preference for basic substrates, and accepts substrates with Arg and Lys in P1 and does not require Ca(2+) for activity. Collectively, these data provide insights into the mechanism and activity of PmC11 and a detailed framework for studies on C11 peptidases from other phylogenetic kingdoms.
Organizational Affiliation:
From the Wellcome Trust Centre for Molecular Parasitology, Institute of Infection, Immunity and Inflammation, College of Medical, Veterinary and Life Sciences, University of Glasgow, Glasgow G12 8TA, United Kingdom.