Beyond heparinization: design of highly potent thrombin inhibitors suitable for surface coupling
Steinmetzer, T., Baum, B., Biela, A., Klebe, G., Nowak, G., Bucha, E.(2012) ChemMedChem 7: 1965-1973
- PubMed: 22907907
- DOI: https://doi.org/10.1002/cmdc.201200292
- Primary Citation of Related Structures:
3UTU - PubMed Abstract:
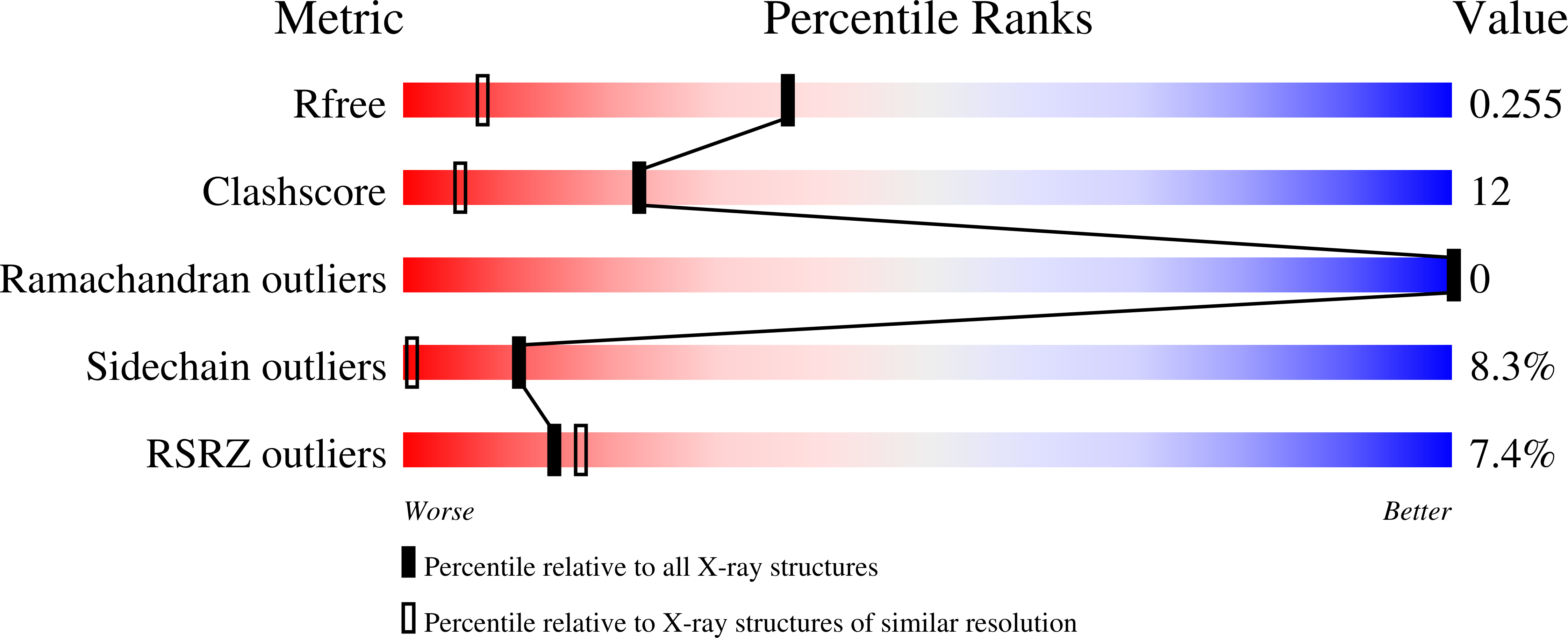
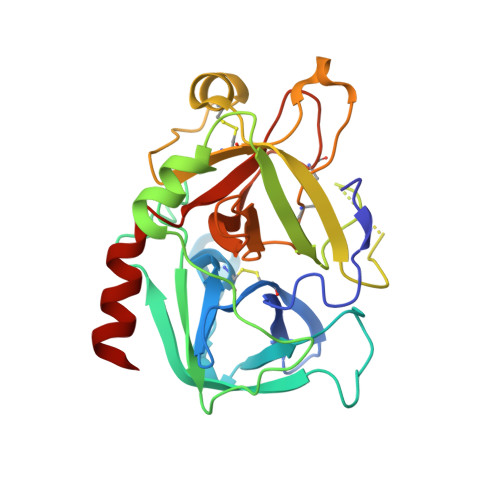
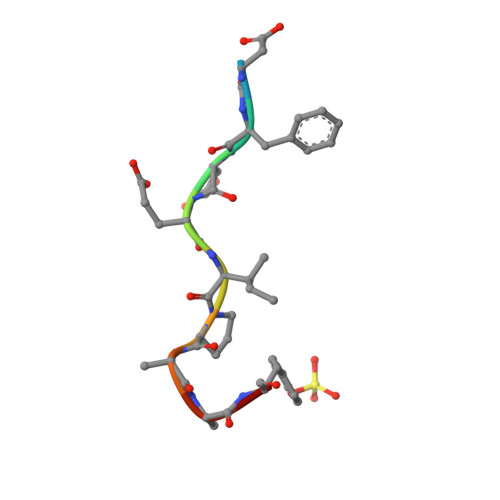
During extracorporeal circulation, when blood comes in contact with artificial surfaces, patients receive a standard treatment with anticoagulants to avoid blood coagulation. Dialysis patients in particular are systemically treated with heparin up to four times a week, causing a high burden for the body. For potential anticoagulant modification of external materials, such as dialysis equipment, a series of highly potent thrombin inhibitors was developed. All inhibitors share the general formula arylsulfonyl-P3-Pro-4-amidinobenzylamide, where P3 is glycyl or a trifunctional amino acid residue in L-configuration. Among this series, several derivatives inhibit thrombin with Ki values of less than 1 nM. Specificity measurements revealed that this inhibitor type is highly specific for thrombin with negligible activity against related trypsin-like serine proteases. X-ray analysis of the most potent analogue in complex with thrombin demonstrated that the N-terminal arylsulfonyl group occupies the aryl binding site, whereas the P3 side chain is directed into the solvent and therefore is well suited for further coupling. Based on their in vitro profile, these inhibitors are suitable candidates for the development of hemocompatible materials with anticoagulant properties.
Organizational Affiliation:
Institute of Pharmaceutical Chemistry, Philipps University, Marbacher Weg 6, 35032 Marburg (Germany). steinmetzer@staff.uni-marburg.de.