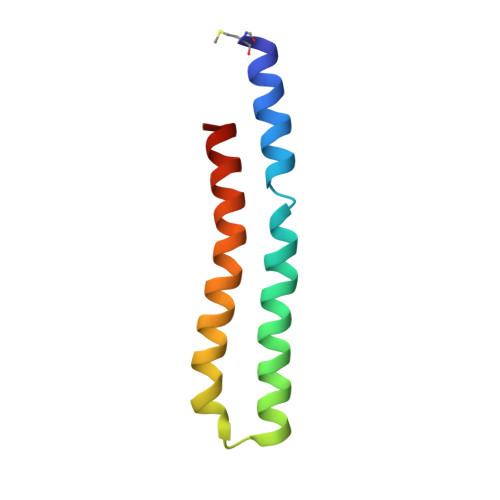
Structure of the c(10) ring of the yeast mitochondrial ATP synthase in the open conformation.
Symersky, J., Pagadala, V., Osowski, D., Krah, A., Meier, T., Faraldo-Gomez, J.D., Mueller, D.M.(2012) Nat Struct Mol Biol 19: 485-491
- PubMed: 22504883
- DOI: https://doi.org/10.1038/nsmb.2284
- Primary Citation of Related Structures:
3U2F, 3U2Y, 3U32, 3UD0 - PubMed Abstract:
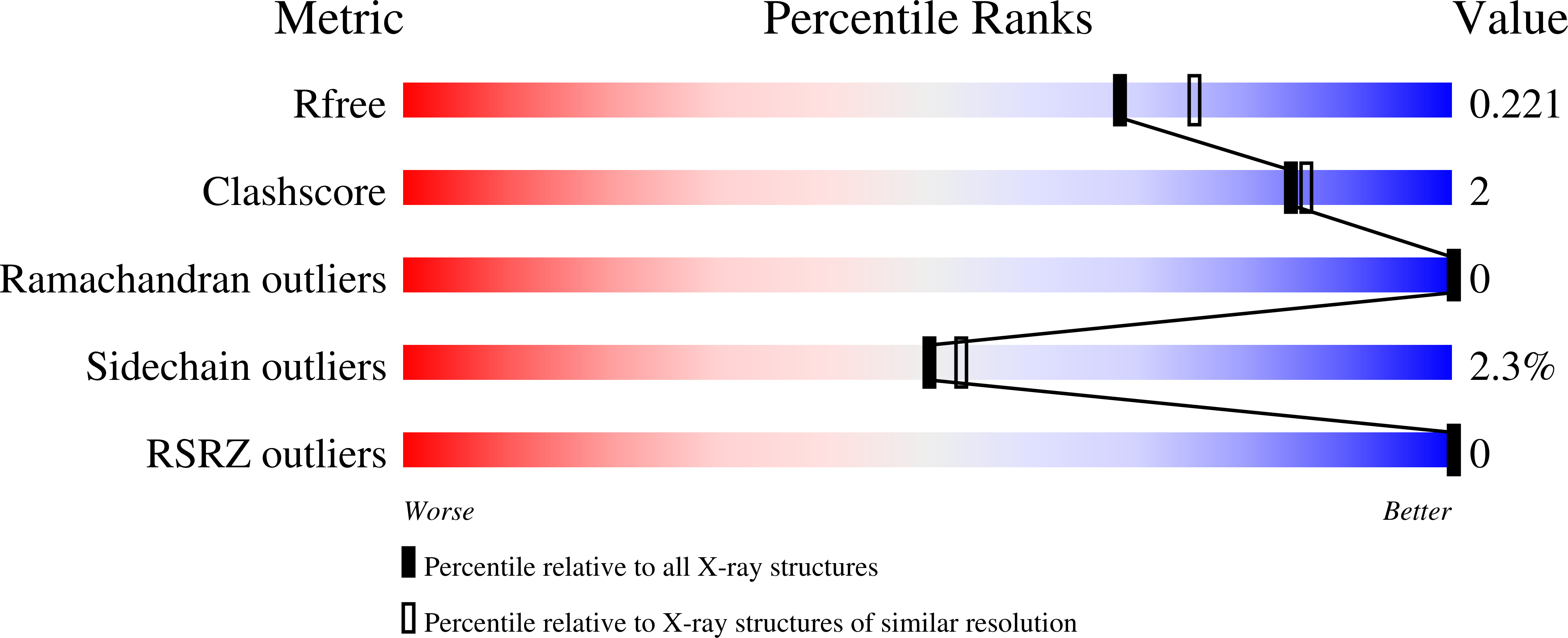
The proton pore of the F(1)F(o) ATP synthase consists of a ring of c subunits, which rotates, driven by downhill proton diffusion across the membrane. An essential carboxylate side chain in each subunit provides a proton-binding site. In all the structures of c-rings reported to date, these sites are in a closed, ion-locked state. Structures are here presented of the c(10) ring from Saccharomyces cerevisiae determined at pH 8.3, 6.1 and 5.5, at resolutions of 2.0 Å, 2.5 Å and 2.0 Å, respectively. The overall structure of this mitochondrial c-ring is similar to known homologs, except that the essential carboxylate, Glu59, adopts an open extended conformation. Molecular dynamics simulations reveal that opening of the essential carboxylate is a consequence of the amphiphilic nature of the crystallization buffer. We propose that this new structure represents the functionally open form of the c subunit, which facilitates proton loading and release.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, Rosalind Franklin University of Medicine and Science, The Chicago Medical School, Chicago, Illinois, USA.