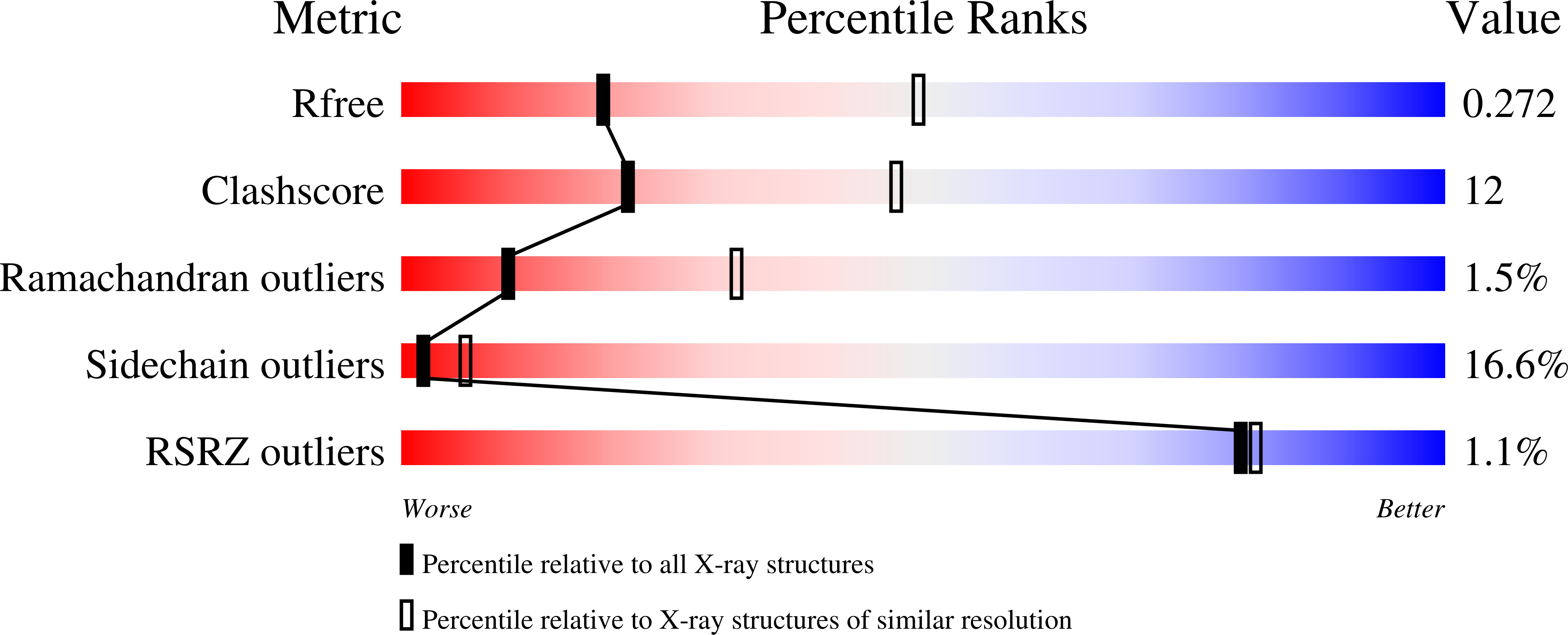
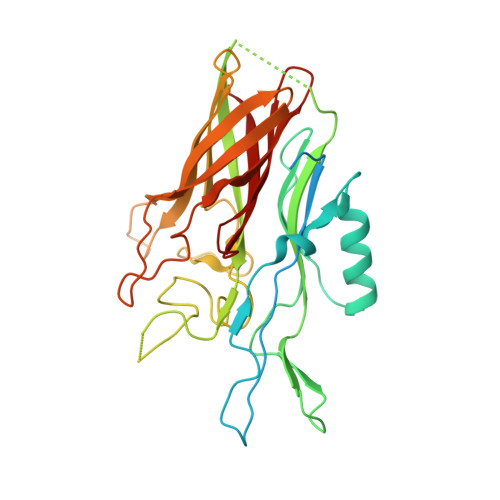
Crystal structure of the N-lobe of lactoferrin binding protein B from Moraxella bovis(1).
Arutyunova, E., Brooks, C.L., Beddek, A., Mak, M.W., Schryvers, A.B., Lemieux, M.J.(2012) Biochem Cell Biol 90: 351-361
- PubMed: 22332934
- DOI: https://doi.org/10.1139/o11-078
- Primary Citation of Related Structures:
3UAQ - PubMed Abstract:
Lactoferrin (Lf) is a bi-lobed, iron-binding protein found on mucosal surfaces and at sites of inflammation. Gram-negative pathogens from the Neisseriaceae and Moraxellaceae families are capable of using Lf as a source of iron for growth through a process mediated by a bacterial surface receptor that directly binds host Lf. This receptor consists of an integral outer membrane protein, lactoferrin binding protein A (LbpA), and a surface lipoprotein, lactoferrin binding protein B (LbpB). The N-lobe of the homologous transferrin binding protein B, TbpB, has been shown to facilitate transferrin binding in the process of iron acquisition. Currently there is little known about the role of LbpB in iron acquisition or how Lf interacts with the bacterial receptor proteins. No structural information on any LbpB or domain is available. In this study, we express and purify from Escherichia coli the full-length LbpB and the N-lobe of LbpB from the bovine pathogen Moraxella bovis for crystallization trials. We demonstrate that M. bovis LbpB binds to bovine but not human Lf. We also report the crystal structure of the N-terminal lobe of LbpB from M. bovis and compare it with the published structures of TbpB to speculate on the process of Lf mediated iron acquisition.
Organizational Affiliation:
Department of Biochemistry, University of Alberta, Edmonton, AB T6G 2H7, Canada.