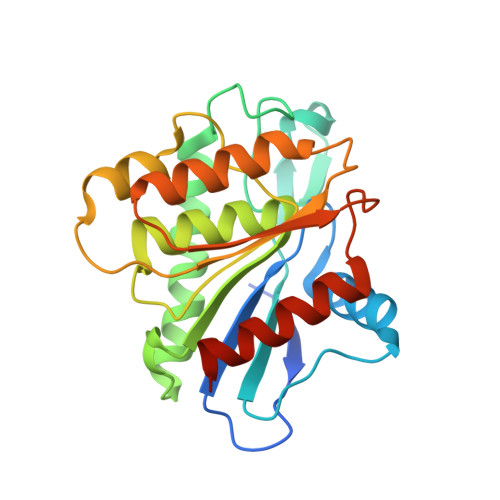
Crystal structure of the predicted phospholipase LYPLAL1 reveals unexpected functional plasticity despite close relationship to acyl protein thioesterases
Burger, M., Zimmermann, T.J., Kondoh, Y., Stege, P., Watanabe, N., Osada, H., Waldmann, H., Vetter, I.R.(2012) J Lipid Res 53: 43-50
- PubMed: 22052940
- DOI: https://doi.org/10.1194/jlr.M019851
- Primary Citation of Related Structures:
3U0V - PubMed Abstract:
Sequence homology indicates the existence of three human cytosolic acyl protein thioesterases, including APT1 that is known to depalmitoylate H- and N-Ras. One of them is the lysophospholipase-like 1 (LYPLAL1) protein that on the one hand is predicted to be closely related to APT1 but on the other hand might also function as a potential triacylglycerol lipase involved in obesity. However, its role remained unclear. The 1.7 Å crystal structure of LYPLAL1 reveals a fold very similar to APT1, as expected, but features a shape of the active site that precludes binding of long-chain substrates. Biochemical data demonstrate that LYPLAL1 exhibits neither phospholipase nor triacylglycerol lipase activity, but rather accepts short-chain substrates. Furthermore, extensive screening efforts using chemical array technique revealed a first small molecule inhibitor of LYPLAL1.
Organizational Affiliation:
Department of Mechanistic Cell Biology, Max Planck Institute of Molecular Physiology, Dortmund, Germany.