Small-molecule binding to the DNA minor groove is mediated by a conserved water cluster.
Wei, D., Wilson, W.D., Neidle, S.(2013) J Am Chem Soc 135: 1369-1377
- PubMed: 23276263
- DOI: https://doi.org/10.1021/ja308952y
- Primary Citation of Related Structures:
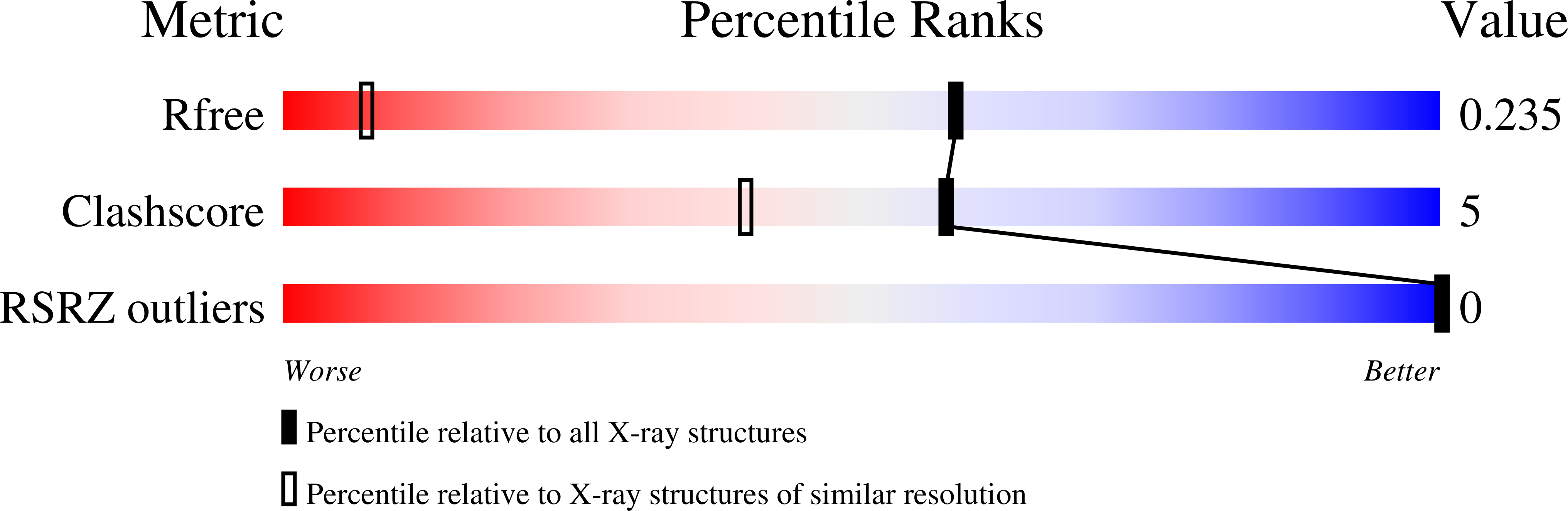
3U05, 3U08, 3U0U, 3U2N - PubMed Abstract:
High-resolution crystal structures of the DNA duplex sequence d(CGCGAATTCGCG)(2) complexed with three minor-groove ligands are reported. A highly conserved cluster of 11 linked water molecules has been found in the native and all 3 ligand-bound structures, positioned at the boundary of the A/T and G/C regions where the minor groove widens. This cluster appears to play a key structural role in stabilizing noncovalently binding small molecules in the AT region of the B-DNA minor groove. The cluster extends from the backbone phosphate groups along the mouth of the groove and links to DNA and ligands by a network of hydrogen bonds that help to maintain the ligands in position. This arrangement of water molecules is distinct from, but linked by, hydrogen bonding to the well-established spine of hydration, which is displaced by bound ligands. Features of the water cluster and observed differences in binding modes help to explain the measured binding affinities and thermodynamic characteristics of these ligands on binding to AT sites in DNA.
Organizational Affiliation:
UCL School of Pharmacy, 29-39 Brunswick Square, London WC1N 1AX, United Kingdom.