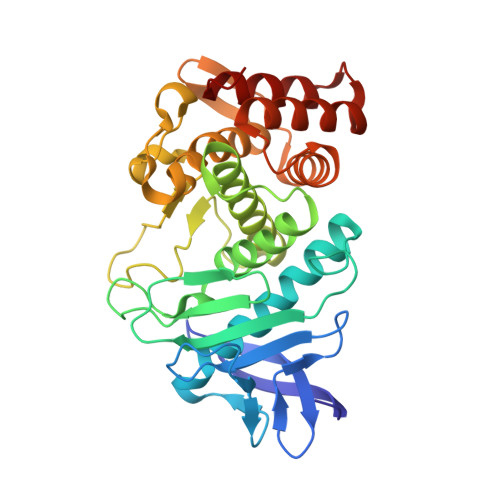
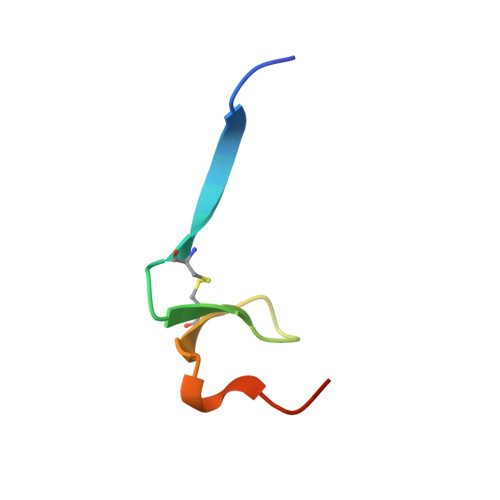
Structural evidence for standard-mechanism inhibition in metallopeptidases from a complex poised to resynthesize a Peptide bond.
Arolas, J.L., Botelho, T.O., Vilcinskas, A., Gomis-Ruth, F.X.(2011) Angew Chem Int Ed Engl 50: 10357-10360
- PubMed: 21915964
- DOI: https://doi.org/10.1002/anie.201103262
- Primary Citation of Related Structures:
3SSB
Organizational Affiliation:
Proteolysis Lab, Molecular Biology Institute of Barcelona, CSIC, Barcelona Science Park, c/Baldiri Reixac, 15-21, 08028 Barcelona, Spain.