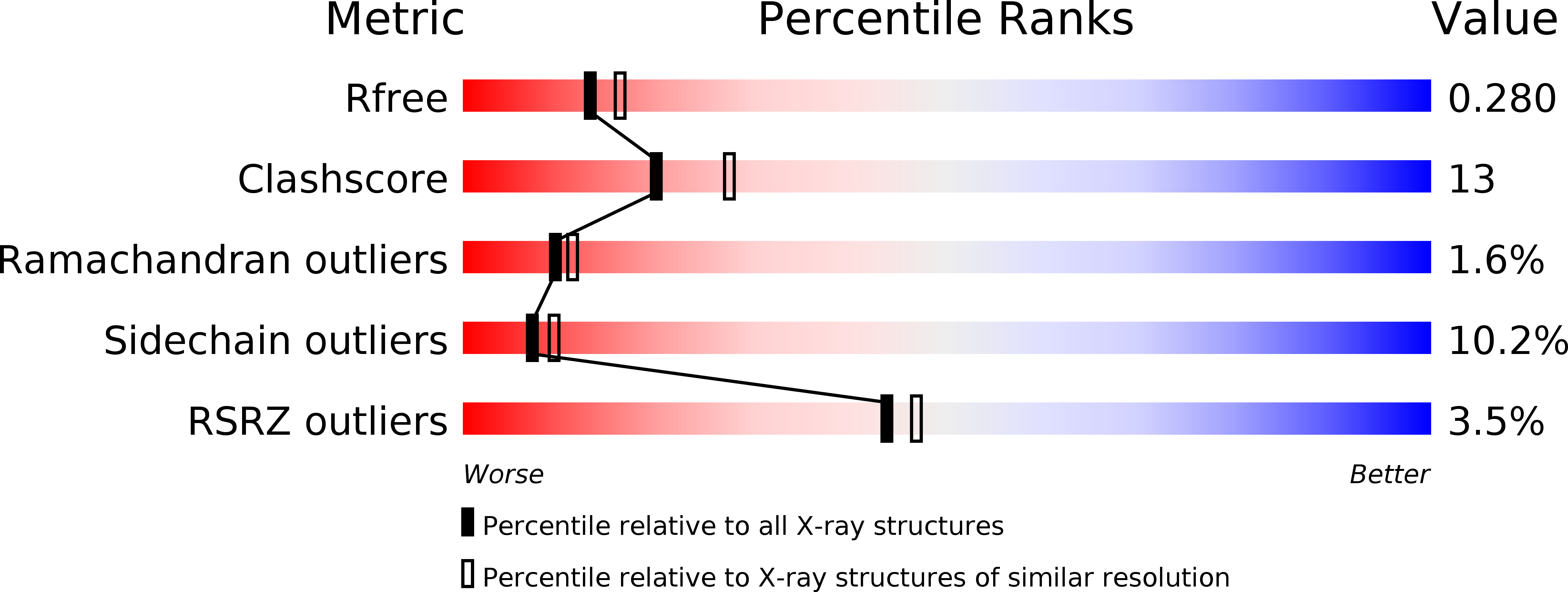
Crystal structure of Staphylococcal dual specific inositol monophosphatase/NADP(H) phosphatase (SAS2203) delineates the molecular basis of substrate specificity
Bhattacharyya, S., Dutta, D., Saha, B., Ghosh, A.K., Das, A.K.(2012) Biochimie 94: 879-890
- PubMed: 22197784
- DOI: https://doi.org/10.1016/j.biochi.2011.12.007
- Primary Citation of Related Structures:
3QMF, 3RYD - PubMed Abstract:
Inositol monophosphatase (IMPase) family of proteins are Mg(2+) activated Li(+) inhibited class of ubiquitous enzymes with promiscuous substrate specificity. Herein, the molecular basis of IMPase substrate specificity is delineated by comparative crystal structural analysis of a Staphylococcal dual specific IMPase/NADP(H) phosphatase (SaIMPase - I) with other IMPases of different substrate compatibility, empowered by in silico docking and Escherichia coli SuhB mutagenesis analysis. Unlike its eubacterial and eukaryotic NADP(H) non-hydrolyzing counterparts, the composite structure of SaIMPase - I active site pocket exhibits high structural resemblance with archaeal NADP(H) hydrolyzing dual specific IMPase/FBPase. The large and shallow SaIMPase - I active site cleft efficiently accommodate large incoming substrates like NADP(H), and therefore, justifies the eminent NADP(H) phosphatase activity of SaIMPase - I. Compared to other NADP(H) non-hydrolyzing IMPases, the profound difference in active site topology as well as the unique NADP(H) recognition capability of SaIMPase - I stems from the differential length and orientation of a distant helix α4 (in human and bovine α5) and its preceding loop. We identified the length of α4 and its preceding loop as the most crucial factor that regulates IMPase substrate specificity by employing a size exclusion mechanism. Hence, in SaIMPase - I, the substrate promiscuity is a gain of function by trimming the length of α4 and its preceding loop, compared to other NADP(H) non-hydrolyzing IMPases. This study thus provides a biochemical - structural framework revealing the length and orientation of α4 and its preceding loop as the predisposing factor for the determination of IMPase substrate specificity.
Organizational Affiliation:
Department of Biotechnology, Indian Institute of Technology Kharagpur, Kharagpur 721302, West Bengal, India.