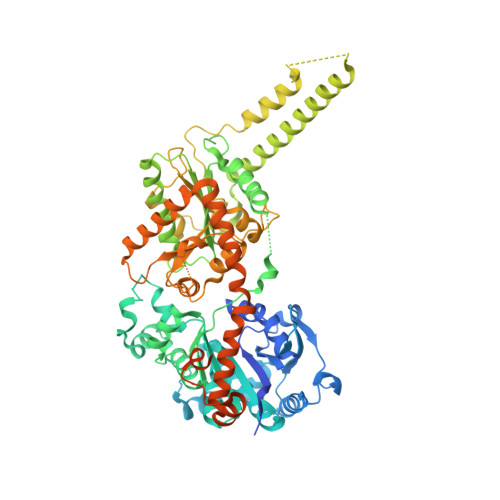
Multiple Glycogen-binding Sites in Eukaryotic Glycogen Synthase Are Required for High Catalytic Efficiency toward Glycogen.
Baskaran, S., Chikwana, V.M., Contreras, C.J., Davis, K.D., Wilson, W.A., Depaoli-Roach, A.A., Roach, P.J., Hurley, T.D.(2011) J Biol Chem 286: 33999-34006
- PubMed: 21835915
- DOI: https://doi.org/10.1074/jbc.M111.264531
- Primary Citation of Related Structures:
3RSZ, 3RT1 - PubMed Abstract:
Glycogen synthase is a rate-limiting enzyme in the biosynthesis of glycogen and has an essential role in glucose homeostasis. The three-dimensional structures of yeast glycogen synthase (Gsy2p) complexed with maltooctaose identified four conserved maltodextrin-binding sites distributed across the surface of the enzyme. Site-1 is positioned on the N-terminal domain, site-2 and site-3 are present on the C-terminal domain, and site-4 is located in an interdomain cleft adjacent to the active site. Mutation of these surface sites decreased glycogen binding and catalytic efficiency toward glycogen. Mutations within site-1 and site-2 reduced the V(max)/S(0.5) for glycogen by 40- and 70-fold, respectively. Combined mutation of site-1 and site-2 decreased the V(max)/S(0.5) for glycogen by >3000-fold. Consistent with the in vitro data, glycogen accumulation in glycogen synthase-deficient yeast cells (Δgsy1-gsy2) transformed with the site-1, site-2, combined site-1/site-2, or site-4 mutant form of Gsy2p was decreased by up to 40-fold. In contrast to the glycogen results, the ability to utilize maltooctaose as an in vitro substrate was unaffected in the site-2 mutant, moderately affected in the site-1 mutant, and almost completely abolished in the site-4 mutant. These data show that the ability to utilize maltooctaose as a substrate can be independent of the ability to utilize glycogen. Our data support the hypothesis that site-1 and site-2 provide a "toehold mechanism," keeping glycogen synthase tightly associated with the glycogen particle, whereas site-4 is more closely associated with positioning of the nonreducing end during catalysis.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, Indiana University School of Medicine, Indianapolis, Indiana 46202, USA.