Crystal Structure of Beta-lactamase/D-alanine Carboxypeptidase from Yersinia pestis complexed with citrate
Kim, Y., Zhou, M., Gu, M., Anderson, W.F., Joachimiak, A., Center for Structural Genomics of Infectious Diseases (CSGID)To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Beta-lactamase/D-alanine Carboxypeptidase | 351 | Yersinia pestis CO92 | Mutation(s): 0 Gene Names: ampH, y2964, YPO1224, YP_0915 | 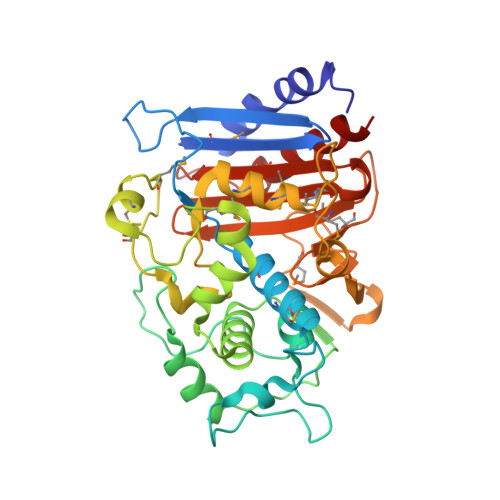 | |
UniProt | |||||
Find proteins for Q0WHI2 (Yersinia pestis) Explore Q0WHI2 Go to UniProtKB: Q0WHI2 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q0WHI2 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| CIT Query on CIT | B [auth A] | CITRIC ACID C6 H8 O7 KRKNYBCHXYNGOX-UHFFFAOYSA-N |  | ||
| GOL Query on GOL | C [auth A], D [auth A], E [auth A] | GLYCEROL C3 H8 O3 PEDCQBHIVMGVHV-UHFFFAOYSA-N |  | ||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| MSE Query on MSE | A | L-PEPTIDE LINKING | C5 H11 N O2 Se |  | MET |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 43.161 | α = 90 |
| b = 75.477 | β = 103.93 |
| c = 59.068 | γ = 90 |
| Software Name | Purpose |
|---|---|
| SBC-Collect | data collection |
| HKL-3000 | data collection |
| HKL-3000 | phasing |
| SHELXS | phasing |
| MLPHARE | phasing |
| BUCCANEER | model building |
| PHENIX | refinement |
| HKL-3000 | data reduction |
| HKL-3000 | data scaling |
| BUCCANEER | phasing |