The crystal structure of glycyl-tRNA synthetase subunit alpha from Campylobacter jejuni subsp. jejuni NCTC in complex with ATP and glycine.
Tan, K., Zhang, R., Zhou, M., Kwon, K., Anderson, W.F., Joachimiak, A.To be published.
Experimental Data Snapshot
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Glycyl-tRNA synthetase alpha subunit | 311 | Campylobacter jejuni | Mutation(s): 0 Gene Names: Cj0704, glyQ EC: 6.1.1.14 | 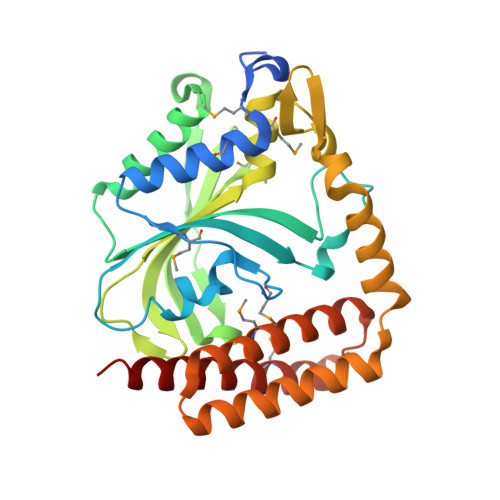 | |
UniProt | |||||
Find proteins for Q9PPK3 (Campylobacter jejuni subsp. jejuni serotype O:2 (strain ATCC 700819 / NCTC 11168)) Explore Q9PPK3 Go to UniProtKB: Q9PPK3 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q9PPK3 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| ATP Query on ATP | D [auth A], E [auth B] | ADENOSINE-5'-TRIPHOSPHATE C10 H16 N5 O13 P3 ZKHQWZAMYRWXGA-KQYNXXCUSA-N |  | ||
| LMR Query on LMR | F [auth B] | (2S)-2-hydroxybutanedioic acid C4 H6 O5 BJEPYKJPYRNKOW-REOHCLBHSA-N |  | ||
| GLY Query on GLY | C [auth A] | GLYCINE C2 H5 N O2 DHMQDGOQFOQNFH-UHFFFAOYSA-N |  | ||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| MSE Query on MSE | A, B | L-PEPTIDE LINKING | C5 H11 N O2 Se |  | MET |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 112.74 | α = 90 |
| b = 112.74 | β = 90 |
| c = 157.753 | γ = 120 |
| Software Name | Purpose |
|---|---|
| SBC-Collect | data collection |
| MOLREP | phasing |
| PHENIX | refinement |
| HKL-3000 | data reduction |
| HKL-3000 | data scaling |