Structural Analysis of the Essential Resuscitation Promoting Factor YeaZ Suggests a Mechanism of Nucleotide Regulation through Dimer Reorganization.
Aydin, I., Saijo-Hamano, Y., Namba, K., Thomas, C., Roujeinikova, A.(2011) PLoS One 6: e23245-e23245
- PubMed: 21858042
- DOI: https://doi.org/10.1371/journal.pone.0023245
- Primary Citation of Related Structures:
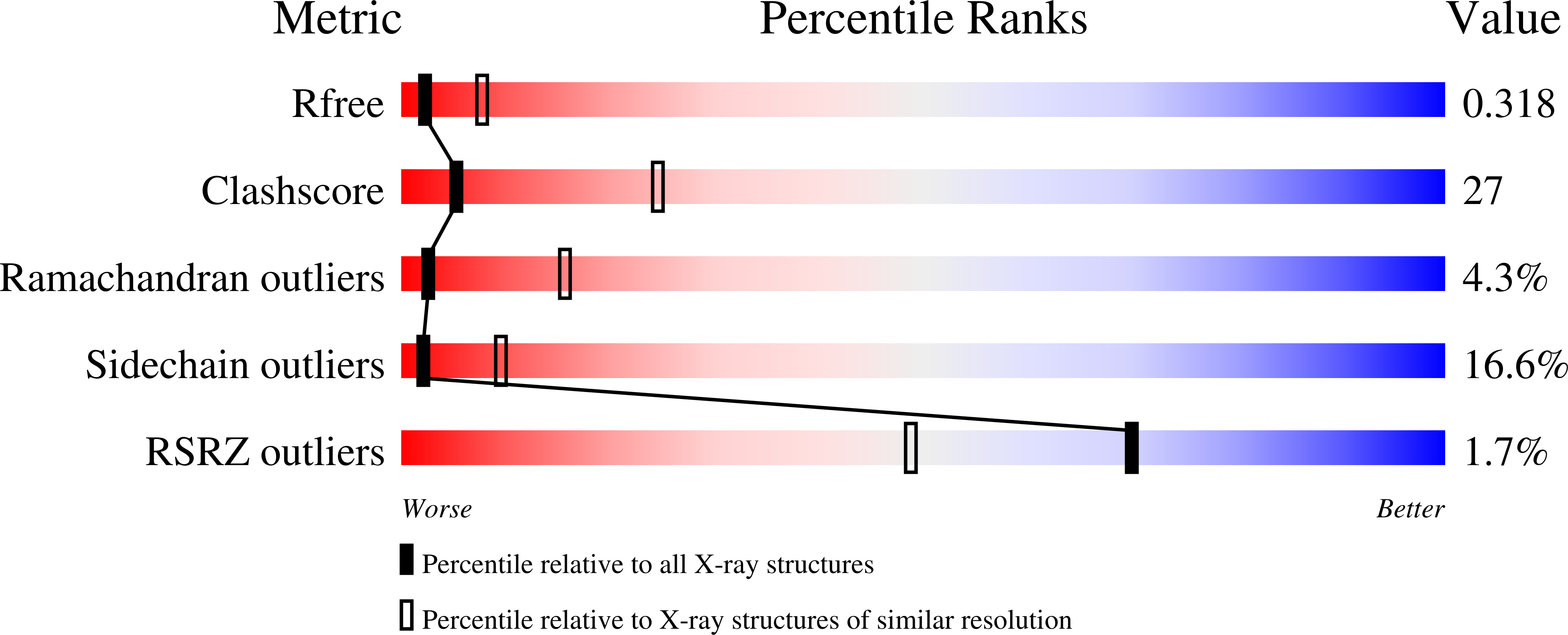
3R6M - PubMed Abstract:
The yeaZ gene product forms part of the conserved network YjeE/YeaZ/YgjD essential for the survival of many gram-negative eubacteria. Among other as yet unidentified roles, YeaZ functions as a resuscitation promoting factor required for survival and resuscitation of cells in a viable but non-culturable (VBNC) state. In order to investigate in detail the structure/function relationship of this family of proteins we have performed X-ray crystallographic studies of Vibrio parahaemolyticus YeaZ. The YeaZ structure showed that it has a classic actin-like nucleotide-binding fold. Comparisons of this crystal structure to that of available homologues from E. coli, T. maritima and S. typhimurium revealed two distinctly different modes of dimer formation. In one form, prevalent in the absence of nucleotide, the putative nucleotide-binding site is incomplete, lacking a binding pocket for a nucleotide base. In the second form, residues from the second subunit complete the nucleotide-binding site. This suggests that the two dimer architectures observed in the crystal structures correspond to a free and a nucleotide-bound form of YeaZ. A multiple sequence alignment of YeaZ proteins from different bacteria allowed us to identify a large conserved hydrophobic patch on the protein surface that becomes exposed upon nucleotide-driven dimer re-arrangement. We hypothesize that the transition between two dimer architectures represents the transition between the 'on' and 'off' states of YeaZ. The effect of this transition is to alternately expose and bury a docking site for the partner protein YgjD. This paper provides the first structural insight into the putative mechanism of nucleotide regulation of YeaZ through dimer reorganization. Our analysis suggests that nucleotide binding to YeaZ may act as a regulator or switch that changes YeaZ shape, allowing it to switch partners between YjeE and YgjD.
Organizational Affiliation:
Department of Microbiology, Monash University, Clayton, Victoria, Australia.