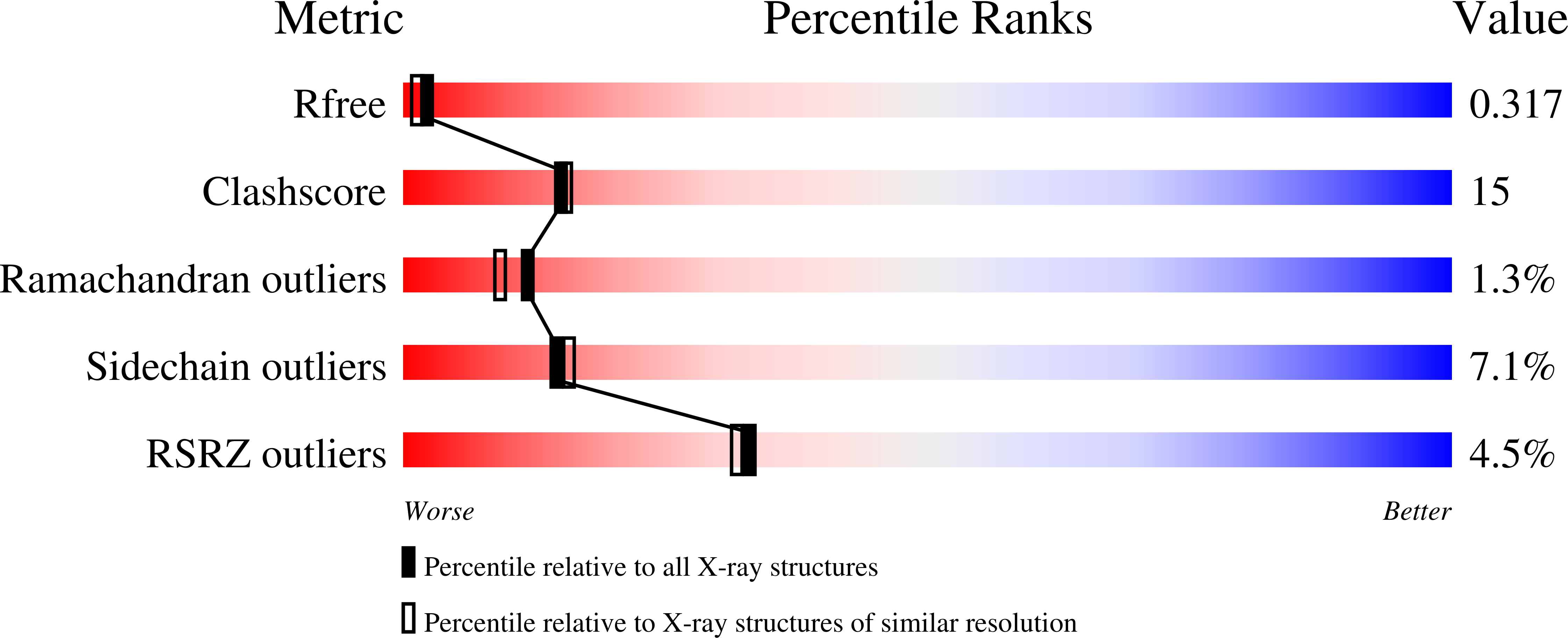
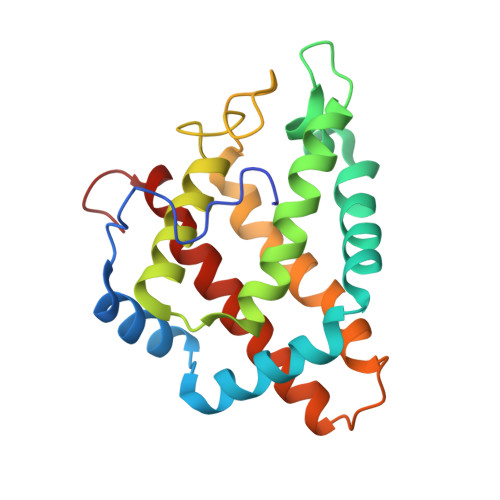
Structural heterogeneity and ligand gating in ferric methanosarcina acetivorans protoglobin mutants.
Pesce, A., Tilleman, L., Dewilde, S., Ascenzi, P., Coletta, M., Ciaccio, C., Bruno, S., Moens, L., Bolognesi, M., Nardini, M.(2011) IUBMB Life 63: 287-294
- PubMed: 21618401
- DOI: https://doi.org/10.1002/iub.484
- Primary Citation of Related Structures:
3QZX, 3QZZ, 3R0G - PubMed Abstract:
Protoglobin from Methanosarcina acetivorans C2A (MaPgb), a strictly anaerobic methanogenic Archaea, displays peculiar structural and functional properties within members of the hemoglobin superfamily. In fact, MaPgb-specific loops and a N-terminal extension (20 amino acid residues) completely bury the heme within the protein matrix. Therefore, the access of diatomic gaseous molecules to the heme is granted by two apolar tunnels reaching the heme distal site from locations at the B/G and B/E helix interfaces. The presence of two tunnels within the protein matrix could be partly responsible for the slightly biphasic ligand binding behavior. Unusually, MaPgb oxygenation is favored with respect to carbonylation. Here, the crucial role of Tyr(B10)61 and Ile(G11)149 residues, located in the heme distal site and lining the protein matrix tunnels 1 and 2, respectively, on ligand binding to the heme-Fe-atom and on distal site structural organization is reported. In particular, tunnel 1 accessibility is modulated by a complex reorganization of the Trp(B9)60 and Phe(E11)93 side-chains, triggered by mutations of the Tyr(B10)61 and Ile(G11)149 residues, and affected by the presence and type of the distal heme-bound ligand.
Organizational Affiliation:
Department of Physics, University of Genova, Genova, Italy.