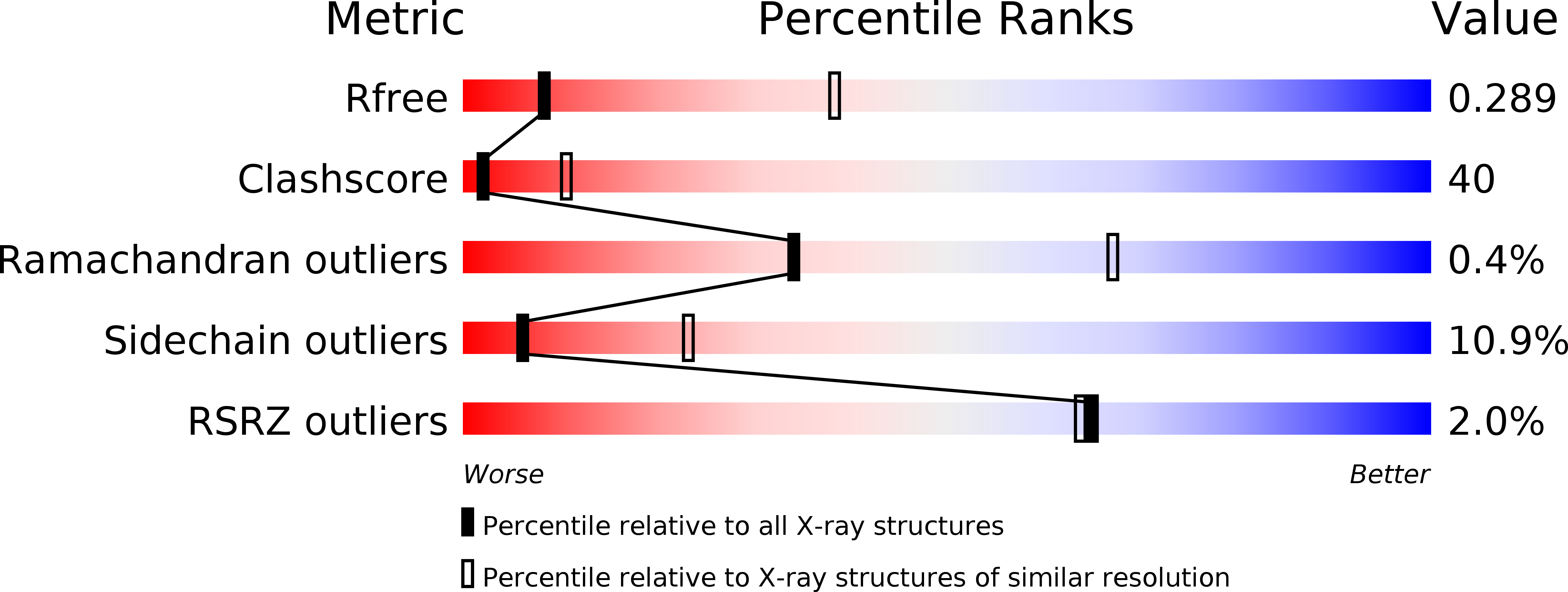
The extracellular architecture of adherens junctions revealed by crystal structures of type I cadherins.
Harrison, O.J., Jin, X., Hong, S., Bahna, F., Ahlsen, G., Brasch, J., Wu, Y., Vendome, J., Felsovalyi, K., Hampton, C.M., Troyanovsky, R.B., Ben-Shaul, A., Frank, J., Troyanovsky, S.M., Shapiro, L., Honig, B.(2011) Structure 19: 244-256
- PubMed: 21300292
- DOI: https://doi.org/10.1016/j.str.2010.11.016
- Primary Citation of Related Structures:
3Q2L, 3Q2N, 3Q2V, 3Q2W - PubMed Abstract:
Adherens junctions, which play a central role in intercellular adhesion, comprise clusters of type I classical cadherins that bind via extracellular domains extended from opposing cell surfaces. We show that a molecular layer seen in crystal structures of E- and N-cadherin ectodomains reported here and in a previous C-cadherin structure corresponds to the extracellular architecture of adherens junctions. In all three ectodomain crystals, cadherins dimerize through a trans adhesive interface and are connected by a second, cis, interface. Assemblies formed by E-cadherin ectodomains coated on liposomes also appear to adopt this structure. Fluorescent imaging of junctions formed from wild-type and mutant E-cadherins in cultured cells confirm conclusions derived from structural evidence. Mutations that interfere with the trans interface ablate adhesion, whereas cis interface mutations disrupt stable junction formation. Our observations are consistent with a model for junction assembly involving strong trans and weak cis interactions localized in the ectodomain.
Organizational Affiliation:
Department of Biochemistry and Molecular Biophysics, Columbia University, New York, NY, USA.