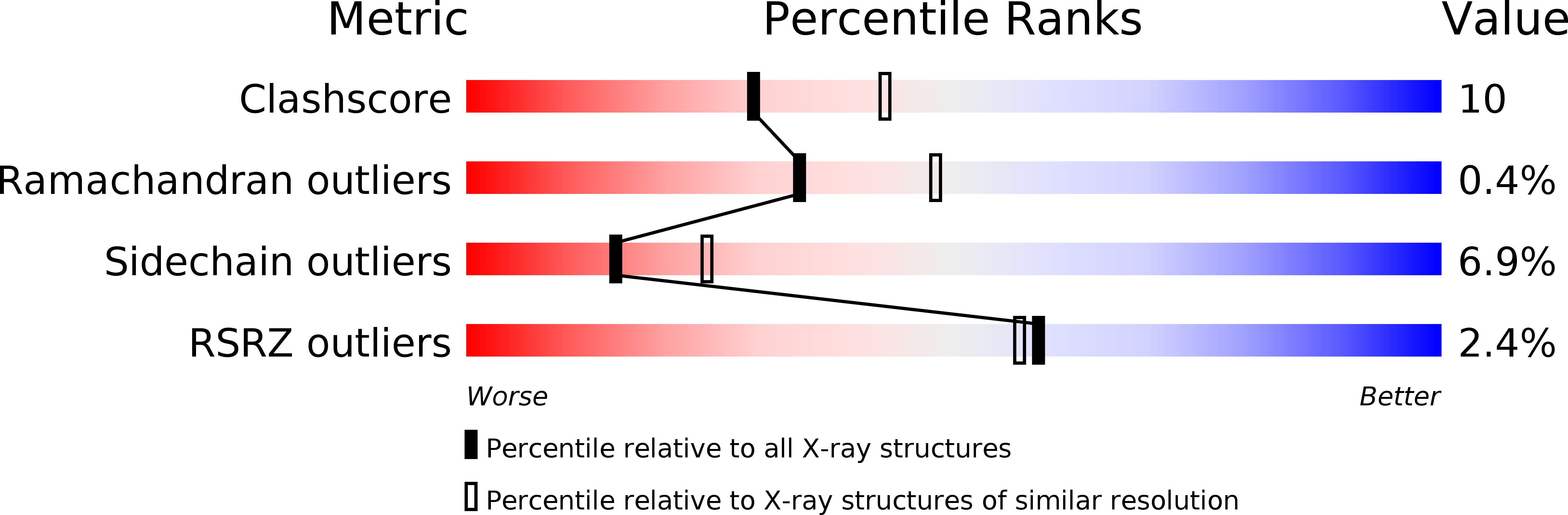
Structure of rabbit muscle phosphoglucomutase refined at 2.4 A resolution.
Liu, Y., Ray, W.J., Baranidharan, S.(1997) Acta Crystallogr D Biol Crystallogr 53: 392-405
- PubMed: 15299905
- DOI: https://doi.org/10.1107/S0907444997000875
- Primary Citation of Related Structures:
1LXT, 3PMG - PubMed Abstract:
Data between 6.0 and 2.4 A resolution, collected at 253 K, wer used to refine a revised atomic model of muscle phosphoglucomutase: final crystallographic R factor = 16.3% (Rfree = 19.1%); final r.m.s. deviations from ideal bond lengths and angles = 0.018 A and 3.2 degrees, respectively. Features of the protein that were recognized only in the revised model include: the disposition of water molecules within domain-domain interfaces; two ion pairs buried in domain-domain interfaces, one of which is a structural arginine around which the active-site phosphoserine loop is wound; the basic architecture of the active-site 'crevice', which is a groove in a 1(1/3)-turn helix, open at both ends, that is produced by the interfacing of the four domains; the distorted hexacoordinate ligand sphere of the active-site Mg2+, where the enzymic phosphate group acts as a bidentate ligand; a pair of arginine residues in domain IV that form part of the enzymic phosphate-binding site (distal subsite) whose disposition in the two monomers of the asymmetric unit is affected unequally by distant crystallographic contacts; structural differences throughout domain IV, produced by these differing contacts, that may mimic solution differences induced by substrate binding; large differences in individually refined Debye-Waller thermal factors for corresponding main-chain atoms in monomers (1) and (2), suggesting a dynamic disorder within the crystal that may involve domain-size groups of residues; and a 'nucleophilic elbow' in the active site that resides in a topological environment differing from previous descriptions of this type of structure in other proteins.
Organizational Affiliation:
The Department of Biological Sciences, Purdue University, West Lafayette, IN 47907, USA.