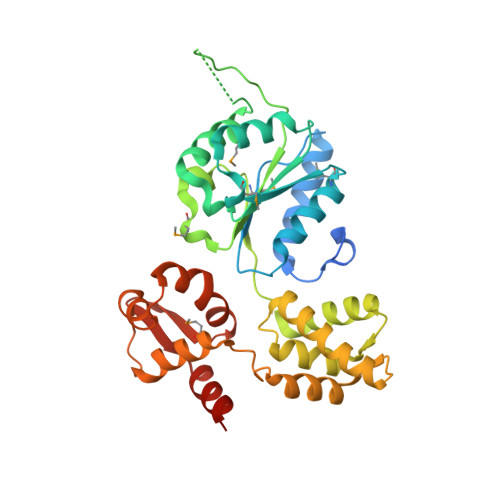
2.7 Angstrom resolution crystal structure of a probable holliday junction DNA helicase (ruvB) from Campylobacter jejuni subsp. jejuni NCTC 11168 in complex with adenosine-5'-diphosphate
Halavaty, A.S., Wawrzak, Z., Skarina, T., Onopriyenko, O., Edwards, A., Savchenko, A., Anderson, W.F., Center for Structural Genomics of Infectious Diseases (CSGID)To be published.