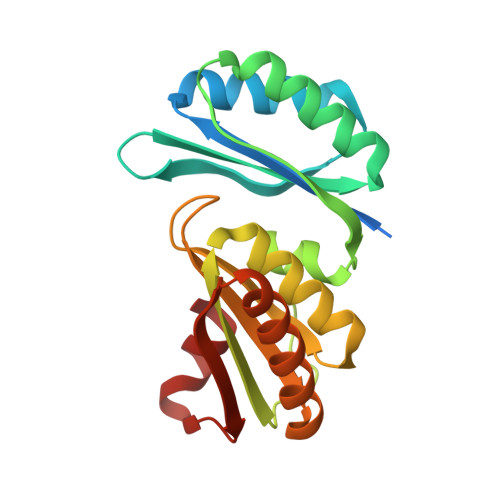
Structure of PduT, a trimeric bacterial microcompartment protein with a 4Fe-4S cluster-binding site
Pang, A., Warren, M.J., Pickersgill, R.W.(2011) Acta Crystallogr D Biol Crystallogr 67: 91-96
- PubMed: 21245529
- DOI: https://doi.org/10.1107/S0907444910050201
- Primary Citation of Related Structures:
3PAC - PubMed Abstract:
Propanediol metabolism in Citrobacter freundii occurs within a metabolosome, a subcellular proteinaceous bacterial microcompartment. The propanediol-utilization (Pdu) microcompartment shell is constructed from thousands of hexagonal-shaped protein complexes made from seven different types of protein subunit. Here, the structure of the bacterial microcompartment protein PduT, which has a tandem structural repeat within the subunit and forms trimers with pseudo-hexagonal symmetry, is reported. This trimeric assembly forms a flat approximately hexagonally shaped disc with a central pore that is suitable for a 4Fe-4S cluster. The essentially cubic shaped 4Fe-4S cluster conforms to the threefold symmetry of the trimer with one free iron, the role of which could be to supply electrons to an associated microcompartment enzyme, PduS.
Organizational Affiliation:
School of Biological and Chemical Sciences, Queen Mary University of London, Mile End Road, London E1 4NS, England.