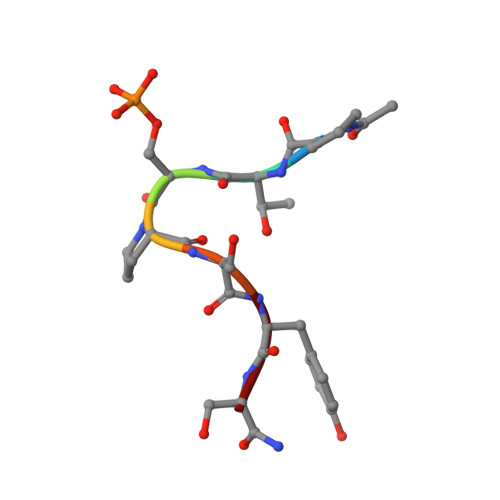
cis-Proline-mediated Ser(P)5 Dephosphorylation by the RNA Polymerase II C-terminal Domain Phosphatase Ssu72.
Werner-Allen, J.W., Lee, C.J., Liu, P., Nicely, N.I., Wang, S., Greenleaf, A.L., Zhou, P.(2011) J Biol Chem 286: 5717-5726
- PubMed: 21159777
- DOI: https://doi.org/10.1074/jbc.M110.197129
- Primary Citation of Related Structures:
3P9Y - PubMed Abstract:
RNA polymerase II coordinates co-transcriptional events by recruiting distinct sets of nuclear factors to specific stages of transcription via changes of phosphorylation patterns along its C-terminal domain (CTD). Although it has become increasingly clear that proline isomerization also helps regulate CTD-associated processes, the molecular basis of its role is unknown. Here, we report the structure of the Ser(P)(5) CTD phosphatase Ssu72 in complex with substrate, revealing a remarkable CTD conformation with the Ser(P)(5)-Pro(6) motif in the cis configuration. We show that the cis-Ser(P)(5)-Pro(6) isomer is the minor population in solution and that Ess1-catalyzed cis-trans-proline isomerization facilitates rapid dephosphorylation by Ssu72, providing an explanation for recently discovered in vivo connections between these enzymes and a revised model for CTD-mediated small nuclear RNA termination. This work presents the first structural evidence of a cis-proline-specific enzyme and an unexpected mechanism of isomer-based regulation of phosphorylation, with broad implications for CTD biology.
Organizational Affiliation:
Department of Biochemistry, Duke University Medical Center, Durham, North Carolina 27710, USA.